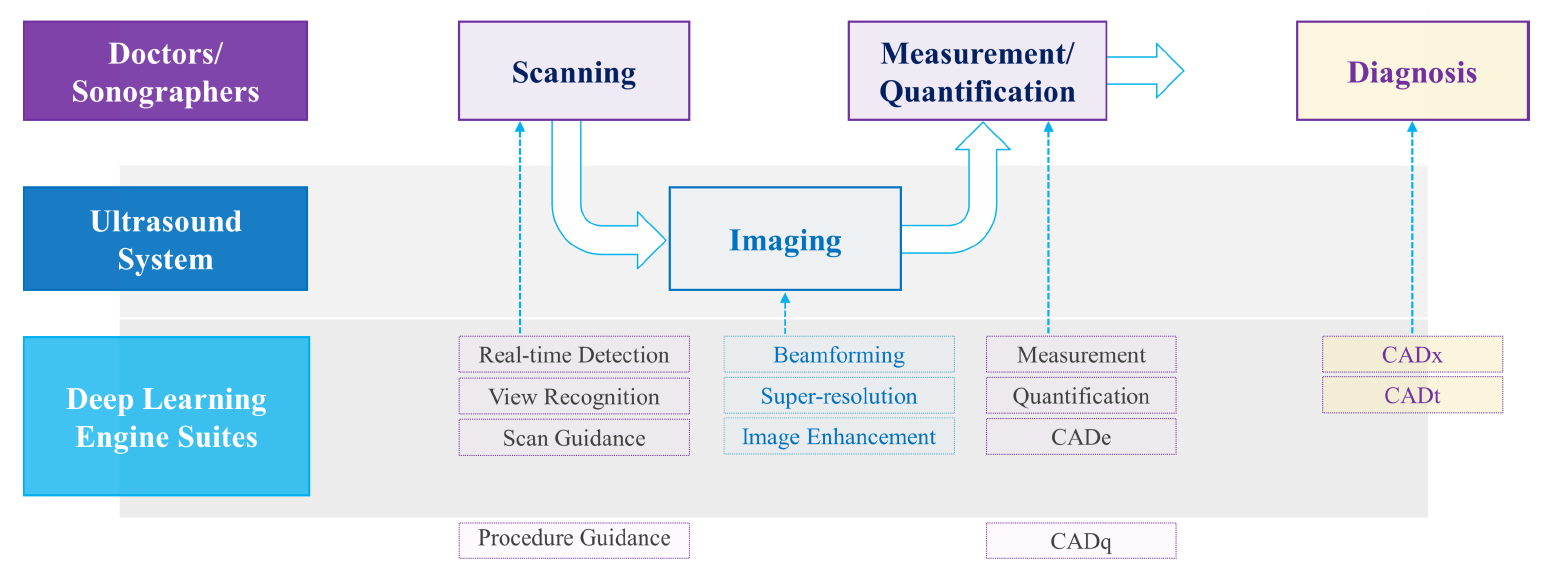
| [1] |
Carneiro G, Nascimento JC. Combining multiple dynamic models and deep learning architectures for tracking the left ventricle endocardium in ultrasound data. IEEE Trans Pattern Anal Mach Intell 2013; 35:2592-2607.
doi: 10.1109/TPAMI.2013.96 |
| [2] |
Carneiro G, Nascimento JC, Freitas A. The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans Image Process 2012; 21:968-982.
doi: 10.1109/TIP.2011.2169273 |
| [3] |
Solomon O, Cohen R, Zhang Y, Yang Y, He Q, Luo J, et al. Deep unfolded robust PCA withapplication to clutter suppression in ultrasound. IEEE Trans Med Imaging 2020; 39:1051-1063.
doi: 10.1109/TMI.42 |
| [4] |
Yoon YH, Khan S, Huh J, Ye JC. Efficient B-mode ultrasound image reconstruction from sub-sampled RF data using deep learning. IEEE Trans Med Imaging 2019; 38:325-336.
doi: 10.1109/TMI.2018.2864821 |
| [5] |
Prevost R, Salehi M, Jagoda S, Kumar N, Sprung J, Ladikos A, et al. 3D freehand ultrasound without external tracking using deep learning. Med Image Anal 2018; 48:187-202.
doi: S1361-8415(18)30371-2 pmid: 29936399 |
| [6] |
Feigin M, Freedman D, Anthony BW. A deep learning framework for single-sided sound speed inversion in medical ultrasound. IEEE Trans Biomed Eng 2020; 67:1142-1151.
doi: 10.1109/TBME.2019.2931195 pmid: 31352330 |
| [7] |
Luchies AC, Byram BC. Training improvements for ultrasound beamforming with deep neural networks. Phys Med Biol 2019; 64:045018.
doi: 10.1088/1361-6560/aafd50 |
| [8] | Yu H, Ding M, Zhang X, Wu J. PCANet based nonlocal means method for speckle noise removal in ultrasound images. PLoS One 2018; 13:e0205390. |
| [9] |
Kim HP, Lee SM, Kwon JY, Park Y, Kim KC, Seo JK. Automatic evaluation of fetal head biometry from ultrasound images using machine learning. Physiol Meas 2019; 40:065009.
doi: 10.1088/1361-6579/ab21ac |
| [10] |
Jafari MH, Girgis H, Van Woudenberg N, Liao Z, Rohling R, Gin K, et al. Automatic biplane left ventricular ejection fraction estimation with mobile point-of-care ultrasound using multi-task learning and adversarial training. Int J Comput Assist Radiol Surg 2019; 14:1027-1037.
doi: 10.1007/s11548-019-01954-w pmid: 30941679 |
| [11] |
Buda M, Wildman-Tobriner B, Castor K, Hoang JK, Mazurowski MA. Deep learning-based segmentation of nodules in thyroid ultrasound: improving performance by utilizing markers present in the images. Ultrasound Med Biol 2020; 46:415-421.
doi: S0301-5629(19)31523-6 pmid: 31699547 |
| [12] |
Loram I, Siddique A, Sanchez MB, Harding P, Silverdale M, Kobylecki C, et al. Objectiveanalysis of neck muscle boundaries for cervical dystonia using ultrasound imaging and deep learning. IEEE J Biomed Health Inform 2020; 24:1016-1027.
doi: 10.1109/JBHI.6221020 |
| [13] |
Park H, Lee HJ, Kim HG, Ro YM, Shin D, Lee SR, et al. Endometrium segmentation ontransvaginal ultrasound image using key-point discriminator. Med Phys 2019; 46:3974-3984.
doi: 10.1002/mp.v46.9 |
| [14] |
Ryou H, Yaqub M, Cavallaro A, Papageorghiou AT, Alison Noble J. Automated 3D ultrasound image analysis for first trimester assessment of fetal health. Phys Med Biol 2019; 64:185010.
doi: 10.1088/1361-6560/ab3ad1 |
| [15] |
Yap MH, Pons G, Marti J, Ganau S, Sentis M, Zwiggelaar R, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J Biomed Health Inform 2018; 22:1218-1226.
doi: 10.1109/JBHI.2017.2731873 |
| [16] | Yap MH, Goyal M, Osman FM, Marti R, Denton E, Juette A, et al. Breast ultrasound lesions recognition: end-to-end deep learning approaches. J Med Imaging (Bellingham) 2019; 6:011007. |
| [17] | Yin S, Peng Q, Li H, Zhang Z, You X, Liu H, et al. Multi-instance deep learning with graph convolutional neural networks for diagnosis of kidney diseases using ultrasound imaging. In: Greenspan H, Tanno R, Erdt M, eds. Uncertainty for safe utilization of machine learning in medical imaging and clinical image-based procedures. Lecture notes in computer science, No. 11840. Cham:Springer, 2019:146-154. |
| [18] |
Lei Y, Tian S, He X, Wang T, Wang B, Patel P, et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net. Med Phys 2019; 46:3194-3206.
doi: 10.1002/mp.13577 pmid: 31074513 |
| [19] |
Van Sloun RJ, Cohen R, Eldar YC. Deep learning in ultrasound imaging. Proc IEEE 2020; 108:11-29.
doi: 10.1109/PROC.5 |
| [20] | Akkus Z, Cai J, Boonrod A, Zeinoddini A, Weston AD, Philbrick KA, et al. A survey of deep-learning applications in ultrasound: artificial intelligence-powered ultrasound for improving clinical workflow. J Am Coll Radiol 2019 ;16(<W>9 Pt B):1318-1328. |
| [21] |
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521:436-444.
doi: 10.1038/nature14539 |
| [22] | Caffe: a deep learning framework [Internet]. Berkeley, CA: University of California, 2020. [cited 2020 Aug 30]. Available from: http://caffe.berkeleyvision.org/. |
| [23] | TensorFlow: TensorFlow is an end-to-end open source platform for machine learning [Internet]. Mountain View, CA: Google, 2020. [cited 2020 Aug 30]. Available from: https://www.tensorflow.org/. |
| [24] | Keras: the high-level API of TensorFlow 2.0 [Internet]. San Francisco, CA: GitHub, 2020. [cited 2020 Aug 30]. Available from: https://keras.io/. |
| [25] | Torch: a scientific computing framework [Internet]. San Francisco, CA: GitHub, 2017. [cited 2020 Aug 30]. Available from: http://torch.ch/. |
| [26] | MXNet: A deep learning framework designed for both efficiency and flexibility [Internet]. San Francisco, CA: GitHub, 2020. [cited 2020 Aug 30]. Available from: https://mxnet.apache.org/. |
| [27] | Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 2012; 25:1097-1105. |
| [28] | Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Preprint at https://arxiv.org/abs/1409.1556 (2015). |
| [29] | Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2015; 2015:1-9. |
| [30] | He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2016; 2016:770-778. |
| [31] | Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2017; 2017:4700-4708. |
| [32] | Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. Proc IEEE Comput Soc Conf Comput Vis Pattern Recogni 2014; 2014:580-587. |
| [33] | Redmon J, Farhadi A. YOLO9000: better, faster, stronger. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2017; 2017:7263-7271. |
| [34] | Redmon J, Farhadi A. YOLOv3: an incremental improvement. Preprint at https://arxiv.org/abs/1804.02767 (2018). |
| [35] | Liu W, Anguelov A, Erhan D, Szegedy C, Reed S, Fu C, et al. SSD:single shot multibox detector. In: Leibe B, Matas J, Sebe N, Welling M, eds. Computer Vision - ECCV 2016. Lecture notes incomputer science, Vol. 9905. Cham: Springer, 2016:12-37. |
| [36] | Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Med Image Comput Comput Assist Interv 2015; 9351:234-241. |
| [37] | Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2015; 2015:3431-3440. |
| [38] | Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. IEEE Int Conf Comput Vis Workshops 2015; 2015:1520-1528. |
| [39] | Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2015; 2015:3431-3440. |
| [40] | Kingma DP, Welling M. Auto-encoding variational bayes. Preprint at (2014). |
| [41] | Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol PA. Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J Mach Learn Res 2010; 11:3371-3408. |
| [42] | Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. Preprint at https://arxiv.org/abs/1511.06434 (2016). |
| [43] | Ledig C, Theis L, Huszar F, Caballero J, Cunningham A, Acosta A, et al. Photo-realistic single image super-resolution using a generative adversarial network. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit 2017; 2017:4681-4690. |
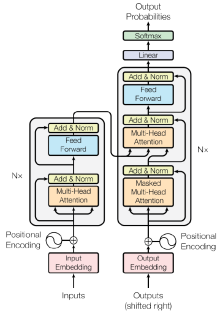
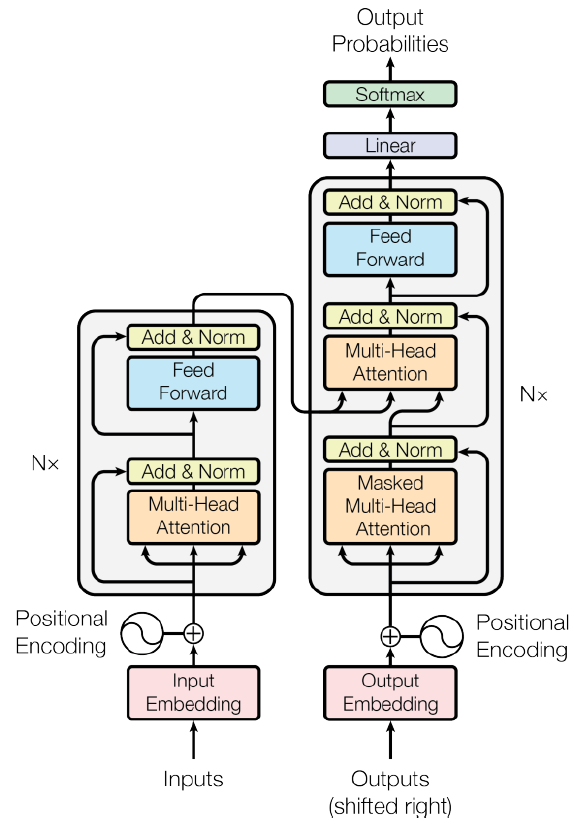
| [44] | Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Advances in neural information processing systems. 2017;30. |
| [45] | Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I. Language models are unsupervised multitask learners. OpenAI blog. 2019 Feb 24;1(8):9. |
| [46] | Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, Neelakantan A, Shyam P, Sastry G, Askell A, Agarwal S. Language models are few-shot learners. Advances in neural information processing systems. 2020; 33:1877-901. |
| [47] |
Yi J, Kang HK, Kwon JH, Kim KS, Park MH, Seong YK, Kim DW, Ahn B, Ha K, Lee J, Hah Z. Technology trends and applications of deep learning in ultrasonography: image quality enhancement, diagnostic support, and improving workflow efficiency. Ultrasonography. 2021 Jan; 40(1):7.
doi: 10.14366/usg.20102 pmid: 33152846 |
| [48] |
Liao WX, He P, Hao J, Wang XY, Yang RL, An D, et al. Automatic identification of breast ultrasound image based on supervised block-based region segmentation algorithm and features combination migration deep learning model. IEEE J Biomed Health Inform 2020; 24:984-993.
doi: 10.1109/JBHI.6221020 |
| [49] |
Tanaka H, Chiu SW, Watanabe T, Kaoku S, Yamaguchi T. Computer-aided diagnosis system for breast ultrasound images using deep learning. Phys Med Biol 2019; 64:235013.
doi: 10.1088/1361-6560/ab5093 |
| [50] |
Zheng X, Yao Z, Huang Y, Yu Y, Wang Y, Liu Y, et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun 2020; 11:1236.
doi: 10.1038/s41467-020-15027-z pmid: 32144248 |
| [51] | Sun Q, Lin X, Zhao Y, Li L, Yan K, Liang D, et al. Deep learning vs. radiomics for predicting axillary lymph node metastasis of breast cancer using ultrasound images: don't forget the peritumoral region. Front Oncol 2020;10-53. |
| [52] |
Nguyen DT, Kang JK, Pham TD, Batchuluun G, Park KR. Ultrasound image-based diagnosis of malignant thyroid nodule using artificial intelligence. Sensors (Basel) 2020; 20:1822.
doi: 10.3390/s20071822 |
| [53] |
Park VY, Han K, Seong YK, Park MH, Kim EK, Moon HJ, et al. Diagnosis of thyroid nodules: performance of a deep learning convolutional neural network model vs. radiologists. Sci Rep 2019; 9:17843.
doi: 10.1038/s41598-019-54434-1 pmid: 31780753 |
| [54] |
Zhu Y, Sang Q, Jia S, Wang Y, Deyer T. Deep neural networks could differentiate Bethesda class III versus class IV/V/VI. Ann Transl Med 2019; 7:231.
doi: 10.21037/atm.2018.07.03 pmid: 31317001 |
| [55] |
Zhang J, Gajjala S, Agrawal P, Tison GH, Hallock LA, Beussink-Nelson L, et al. Fully automated echocardiogram interpretation in clinical practice. Circulation 2018; 138:1623-1635.
doi: 10.1161/CIRCULATIONAHA.118.034338 pmid: 30354459 |
| [56] | Iftikhar P, Kuijpers MV, Khayyat A, Iftikhar A, DeGouvia De Sa M. Artificial intelligence: a newparadigm in obstetrics and gynecology research and clinical practice. Cureus 2020; 12:e7124. |
| [57] |
Yaqub M, Kelly B, Papageorghiou AT, Noble JA. A deep learning solution for automatic fetal neurosonographic diagnostic plane verification using clinical standard constraints. Ultrasound Med Biol 2017; 43:2925-2933.
doi: S0301-5629(17)30332-0 pmid: 28958729 |
| [58] |
Zhang L, Huang J, Liu L. Improved deep learning network based in combination with cost-sensitive learning for early detection of ovarian cancer in color ultrasound detecting system. J Med Syst 2019; 43:251.
doi: 10.1007/s10916-019-1356-8 pmid: 31254110 |
| [59] | Khazendar S, Sayasneh A, Al-Assam H, Du H, Kaijser J, Ferrara L, et al. Automated characterisation of ultrasound images of ovarian tumours: the diagnostic accuracy of a support vector machine and image processing with a local binary pattern operator. Facts Views Vis Obgyn |
| [60] |
Chang RF, Wu WJ, Moon WK, Chen DR. Improvement in breast tumor discrimination by support vector machines and speckle-emphasis texture analysis. Ultrasound Med Biol 2003; 29:679-686.
doi: 10.1016/S0301-5629(02)00788-3 |
| [61] | Attia MW, Abou-Chadi FE, Moustafa HE, Mekky N. Classification of ultrasound kidney images using PCA and neural networks. Int J Adv Comput Sci Appl 2015; 6:53-57. |
| [62] |
Bridge CP, Ioannou C, Noble JA. Automated annotation and quantitative description of ultrasound videos of the fetal heart. Med Image Anal 2017; 36:147-161.
doi: S1361-8415(16)30208-0 pmid: 27907850 |
| [63] |
Hsu WY. Automatic left ventricle recognition, segmentation and tracking in cardiac ultrasound image sequences. IEEE Access 2019; 7:140524-140533.
doi: 10.1109/Access.6287639 |
| [64] | Girshick R. Fast R-CNN. Proc IEEE Int Conf Comput Vis 2015; 2015:1440-1448. |
| [65] |
Cootes TF, Taylor CJ, Cooper DH, Graham J. Active shape models: their training and application. Comput Vis Image Underst 1995; 61:38-59.
doi: 10.1006/cviu.1995.1004 |
| [66] |
Qu R, Xu G, Ding C, Jia W, Sun M. Deep learning-based methodology for recognition of fetal brain standard scan planes in 2D ultrasound images. IEEE Access 2019; 8:44443-44451.
doi: 10.1109/Access.6287639 |
| [67] |
Cai Y, Sharma H, Chatelain P, Noble JA. SonoEyeNet: standardized fetal ultrasound plane detection informed by eye tracking. Proc IEEE Int Symp Biomed Imaging 2018; 2018:1475-1478.
doi: 10.1109/ISBI.2018.8363851 pmid: 30972215 |
| [68] |
Baumgartner CF, Kamnitsas K, Matthew J, Fletcher TP, Smith S, Koch LM, et al. SonoNet: real-time detection and localisation of fetal standard scan planes in freehand ultrasound. IEEE Trans Med Imaging 2017; 36:2204-2215.
doi: 10.1109/TMI.42 |
| [69] | Kwon JY. The intelligent way to evaluate fetal CNS: ViewAssist™ and BiometryAssistTM. White Paper, Samsung Medison. 2022 August 25. |
| [70] | Milletari F, Birodkar V, Sofka M. Straight to the point:reinforcement learning for user guidance in ultrasound. In: Wang Q, Gomez A, Hutter J, McLeod K, Zimmer V, Zettinig O, eds. Smart ultrasound imaging and perinatal, preterm and paediatric image analysis. Cham: Springer, 2019:3-10. |
| [71] | Jarosik P, Lewandowski M. Automatic ultrasound guidance based on deep reinforcement learning. IEEE Int Ultrason Symp 2019; 2019:475-478. |
| [72] | Kwon JY. Feasibility of improved algorithm-based BiometryAssistTM in fetal biometric measurement. White Paper, Samsung Medison. 2020 May 27. |
| [73] |
Rizzo G, Pietrolucci ME, Capponi A, Mappa I. Exploring the role of artificial intelligence in the study of fetal heart. The International Journal of Cardiovascular Imaging. 2022 May; 38(5):1017-9.
doi: 10.1007/s10554-022-02588-x |
| [74] | Chang HJ. HeartAssist™: Automatic View Classification and Measurement Tool for Adult Transthoracic Echocardiography. White Paper, Samsung Medison. 2022 September 14. |
| [75] | Smistad E, Ostvik A, Salte IM, Melichova D, Nguyen TM, Haugaa K, et al. Real-time automatic ejection fraction and foreshortening detection using deep learning. IEEE Trans Ultrason Ferroelectr Freq Control 2020 Mar 16 [Epub]. https://doi.org/10.1109/TUFFC.2020.2981037. |
| [76] |
Sobhaninia Z, Rafiei S, Emami A, Karimi N, Najarian K, Samavi S, et al. Fetal ultrasound image segmentation for measuring biometric parameters using multi-task deep learning. Annu Int Conf IEEE Eng Med Biol Soc 2019; 2019:6545-6548.
doi: 10.1109/EMBC.2019.8856981 pmid: 31947341 |
| [77] | Kwon JY. Feasibility of improved algorithm-based BiometryAssist in fetal biometric measurement(white paper) [Internet]. Seongnam: Samsung Healthcare, 2019. [cited 2020 Aug 30]. Available from: https://www.samsunghealthcare.com/en/knowledge_hub/clinical_library/white_paper. |
| [78] | Jeon SK, Lee JM, Joo I, Yoon JH, Lee G. Two-dimensional convolutional neural network using quantitative US for noninvasive assessment of hepatic steatosis in NAFLD. Radiology. 2023 Jan 3:221510. |
| [79] |
Yaprak M, Cakir O, Turan MN, Dayanan R, Akin S, Degirmen E, et al. Role of ultrasonographic chronic kidney disease score in the assessment of chronic kidney disease. Int Urol Nephrol 2017; 49:123-131.
doi: 10.1007/s11255-016-1443-4 pmid: 27796695 |
| [80] |
Kuo CC, Chang CM, Liu KT, Lin WK, Chiang HY, Chung CW, et al. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. NPJ Digit Med 2019; 2:29.
doi: 10.1038/s41746-019-0104-2 |
| [81] |
Lin BS, Chen JL, Tu YH, Shih YX, Lin YC, Chi WL, et al. Using deep learning in ultrasound imaging of bicipital peritendinous effusion to grade inflammation severity. IEEE J Biomed Health Inform 2020; 24:1037-1045.
doi: 10.1109/JBHI.6221020 |
| [82] |
Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, et al. Deep learning radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut 2019; 68:729-741.
doi: 10.1136/gutjnl-2018-316204 pmid: 29730602 |
| [83] |
Castera L, Vergniol J, Foucher J, Le Bail B, Chanteloup E, Haaser M, et al. Prospective comparison of transient elastography, Fibrotest, APRI, and liver biopsy for the assessment of fibrosis in chronic hepatitis C. Gastroenterology 2005; 128:343-350.
doi: 10.1053/j.gastro.2004.11.018 pmid: 15685546 |
| [84] |
Rousselet MC, Michalak S, Dupre F, Croue A, Bedossa P, Saint-Andre JP, et al. Sources of variability in histological scoring of chronic viral hepatitis. Hepatology 2005; 41:257-264.
doi: 10.1002/hep.20535 |
| [85] |
Xue LY, Jiang ZY, Fu TT, Wang QM, Zhu YL, Dai M, et al. Transfer learning radiomics based on multimodal ultrasound imaging for staging liver fibrosis. Eur Radiol 2020; 30:2973-2983.
doi: 10.1007/s00330-019-06595-w |
| [86] |
Hwang SI, Ahn H, Lee HJ, Hong SK, Byun SS, Lee S, Choe G, Park JS, Son Y. Comparison of Accuracies between Real-Time Nonrigid and Rigid Registration in the MRI-US Fusion Biopsy of the Prostate. Diagnostics. 2021 Aug 16; 11(8):1481.
doi: 10.3390/diagnostics11081481 |
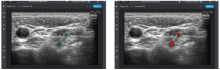
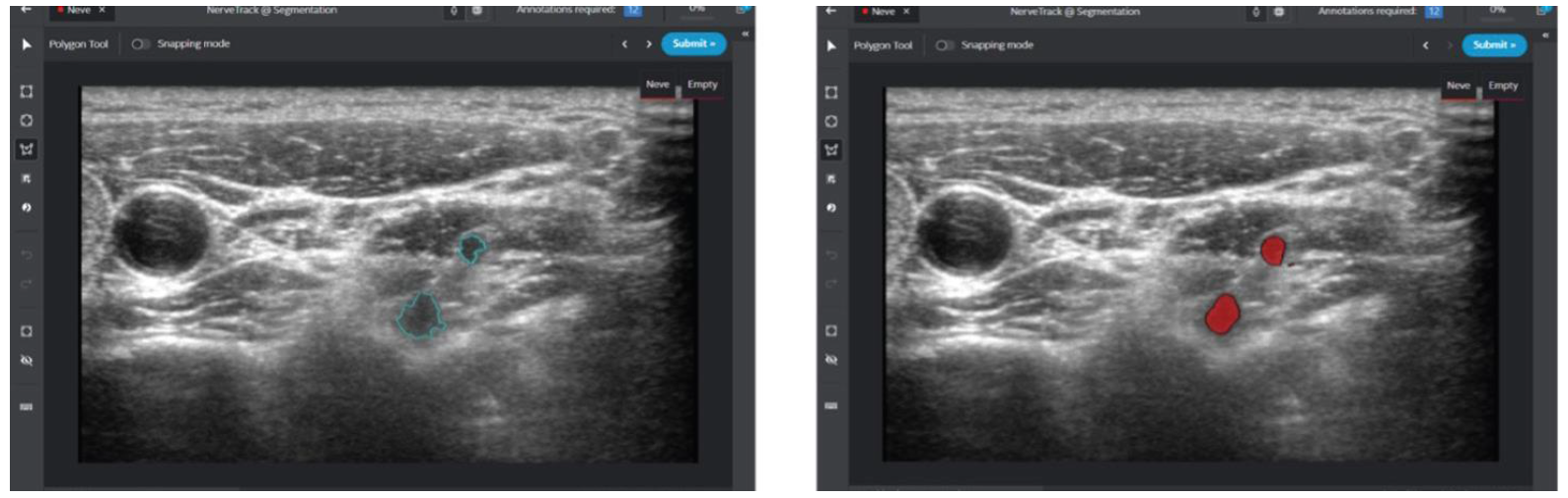
| [87] | Intel GetiTM Platform Accelerates AI Model Training for Real-Time Nerve Detection in Samsung Ultrasound Systems. Case Studies, Intel. 2022 November 24. |
| No related articles found! |
|
||