Advanced Ultrasound in Diagnosis and Therapy ›› 2025, Vol. 9 ›› Issue (4): 388-408.doi: 10.26599/AUDT.2025.250101
Previous Articles Next Articles
Received:2025-10-15
Revised:2025-10-28
Accepted:2025-11-05
Online:2025-12-30
Published:2025-11-06
Contact:
Department of Medical Ultrasonics, Institute of Diagnostic and Interventional Ultrasound, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou 510080, China (Xiaoyan Xie), e-mail: xiexyan@mail.sysu.edu.cn (XY X).,
Zhong Xian, Xie Xiaoyan. Multimodal Ultrasound Radiomics in Liver Disease: Current Status and Future Directions. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 388-408.
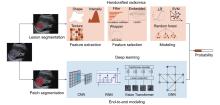
Figure 1
Workflow of handcrafted radiomics and deep learning. LASSO, least absolute shrinkage and selection operator; RFE, recursive feature elimination; LR, logistic regression; SVM, support vector machines; CNN, convolutional neural network. GNN, graph neural network; RNN, recurrent neural networks."
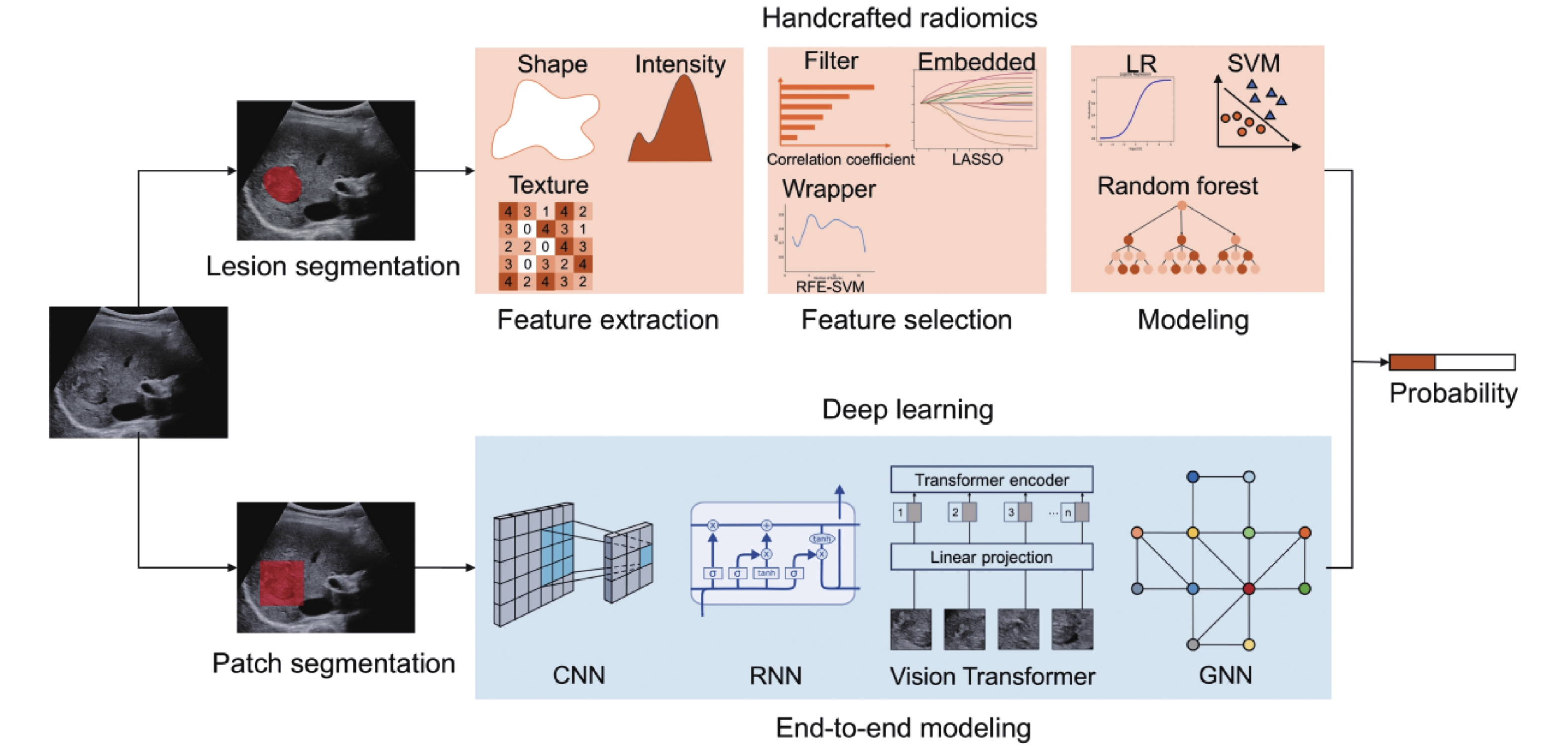
Table 1
Summary of published radiomics studies on fatty liver disease"
| Reference | Study design | Imaging modality | No. of patients/ images | Radiomics technique | Detailed method | Task | Reference standard | Results | |||
| AUC | Accuracy | Sensitivity | Specificity | ||||||||
| HR, handcrafted radiomics; DL, deep learning; FLD, fatty liver disease; CAP, controlled attenuation parameter; CNN, convolutional neural networks; MRI-PDFF, magnetic resonance imaging derived proton density fat fraction; ORF, original radio frequency signal; CDFI, Color doppler flow imaging | |||||||||||
| Wu 2022 [ | Retrospective, single center | B-mode | 321 images from 235 patients | HR | Different classifiers | FLD diagnosis | CAP and biopsy | 0.75 | 70 | 66.4 | NA |
| Cao 2020 [ | Retrospective, single center | B-mode | 240 images from 240 patients | DL | CNN | FLD diagnosis and severity classification | US evaluation by radiologists | 0.933-0.958 | |||
| Byra 2018 [ | Retrospective, single center | B-mode | 550 images from 55 patients | DL | Inception-ResNet-v2+SVM | FLD diagnosis | Liver biopsy | 0.977 | 96.3 | 100 | 88.2 |
| Reddy 2018 [ | Retrospective, single center | B-mode | 157 images | DL | VGG16 Transfer Learning | FLD diagnosis | US evaluation by radiologists | 0.96 | 90.6 | 95 | 85 |
| Kim 2021 [ | Retrospective, single center | B-mode | 180 images from 90 cases | DL | VGG19 | FLD diagnosis | MRI–PDFF | 0.87 | 80.1 | NA | 80.5 |
| Han 2020 [ | Prospective, single center | ORF | 204 patients | DL | One-dimensional CNN | FLD diagnosis and fat fraction estimation | MRI–PDFF | 0.98 | 96 | 97 | 94 |
| Chou 2021 [ | Retrospective, single center | B-mode | 21855 images from 2070 patients | DL | ResNet-50 v2 | FLD diagnosis and severity classification | US evaluation by radiologists | 0.971-0.996 | |||
| Tahmasebi 2023 [ | Prospective, single center | B-mode | 1435 images from 130 patients | DL | Google AutoML | FLD diagnosis | MRI–PDFF | NA | 83.4 | 72.2 | 94.6 |
| Rhyou 2021 [ | Retrospective, multicenter | B-mode | 3200 images from 1902 patients' | DL | Cascaded Neural Network | FLD diagnosis and severity classification | US evaluation by radiologists | NA | 99.6 | 99.1 | 100 |
| Yang 2023 [ | Retrospective, single center | B-mode | 1856 images from 928 patients | DL | Two-section neural network | FLD diagnosis and severity classification | US evaluation by radiologists | 0.84-0.93 | 76.3 | ||
| Liu 2024 [ | Prospective, single center | B-mode +CDFI | 710 images from 710 patients | DL+HR | VGG16 model and two HR features | FLD diagnosis and severity classification | NA | 0.836-0.947 | 77.5 | ||
Table 2
Summary of published ultrasound radiomics studies for diagnosis and grading of liver fibrosis"
| Reference | Study design | Imaging modality | No. of patients/images | Radiomics technique | Detailed method | Task | Reference standard | Results | |||
| AUC | Accuracy | Sensitivity | Specificity | ||||||||
| HR, handcrafted radiomics; DL, deep learning; TE, transient elastography; CNN, convolutional neural networks; GAN, generative adversarial network; ORF, original radio frequency signal; SWE, shear wave elastography; SVM, support vector machines; LSM, liver stiffness measurement; STQ, sound touch quantification; STE, sound touch elastography | |||||||||||
| Meng 2017 [ | Retrospective, single center | B-mode | 279 patients | DL | Transfer Learning and FCNet | Liver fibrosis classification | NA | 93.9 | |||
| Lee 2020 [ | Retrospective, single center | B-mode | 3446 patients | DL | DCNN | Liver fibrosis classification | Pathology or TE | 0.857 for cirrhosis prediction | 88.3 | 77.8 | 93.7 |
| Feng 2021 [ | Retrospective, single center | B-mode | 286 patients | DL | Pyramid-structured CNN | Liver fibrosis classification | Liver biopsy | 0.99 | 95.7 | 96.5 | |
| Ruan 2021 [ | Retrospective, multicenter | B-mode | 508 patients | DL | Multi-scale texture network (MSTNet) | Liver fibrosis classification | Liver biopsy | 0.92 (≥F2) 0.89 (F4) | 85.1 (≥F2) 87.8 (F4) | 87.6 (≥F2) 78.1 (F4) | |
| Duan 2022 [ | Retrospective, multicenter | B-mode | 434 patients | DL+HR | DL features generated by GAN and HR features | Liver fibrosis screening | Liver biopsy | 0.861 for cirrhosis prediction | |||
| Joo 2023 [ | Retrospective, multicenter | B-mode | 955 patients | DL | Transfer learning of different models | Liver fibrosis classification | Liver biopsy | 0.8592 | |||
| Park 2024 [ | Retrospective, multicenter | B-mode | 933 patients | DL | EfficientNet | Liver fibrosis classification | Liver surgery and biopsy | 0.94-0.96 for F0-F4 | 93-96 | 79-89 | 95-98 |
| Ai 2024 [ | Retrospective, single center | ORF | 237 patients | DL | 2D CNN for segmentation and 1D CNN for classification | Liver fibrosis classification | Liver biopsy | 0.957 (≥F1) 0.729 (≥F3) 0.876 (≥F4) | |||
| Zhang 2025 [ | Retrospective, multicenter | high-frequency B-mode images | 1500 patients | DL | InceptionNeXt | Liver fibrosis classification | Liver biopsy | 0.81 (≥S2) 0.93 (≥S3) 0.87 (S4) | 74 (≥S2) 85 (≥S3) 84 (S4) | 84 (≥S2) 85 (≥S3) 79 (S4) | 65 (≥S2) 86 (≥S3) 85 (S4) |
| Chen 2017 [ | Retrospective, multicenter | Real-time tissue elastography | 513 patients | HR | Random Forest | Liver fibrosis classification | Liver biopsy | 82 | 75 | 86 | |
| Gatos 2016 [ | Retrospective, single center | SWE | 85 patients | HR | Color to stiffness mapping and SVM | Liver fibrosis classification | Liver biopsy | 0.85 | 87 | 83.3 | 89.1 |
| Gatos 2017 [ | Retrospective, single center | SWE | 126 patients | HR | Stiffness value clustering and SVM | Liver fibrosis classification | Liver biopsy | 0.87 | 87.3 | 93.5 | 81.2 |
| Gatos 2019 [ | Retrospective, single center | SWE | 200 patients | DL | A wavelet transform and fuzzy c-means clustering algorithm to detect areas of high and low stiffness temporal stability | Liver fibrosis classification | Liver biopsy | 0.89-0.95 | 82.5-92.5 | ||
| Kagadis 2020 [ | Retrospective, single center | SWE | 200 patients | DL | Transfer learning of different models | Liver fibrosis classification | Liver biopsy | 0.979-0.990 | 87.2-97.4 | ||
| Wang 2018 [ | Prospective, multicenter | SWE | 398 patients | DL | Deep learning radiomics of elastography (DLRE) | Liver fibrosis classification | Liver biopsy | 0.85 (≥F2) 0.98 (≥F3) 0.97 (F4) | 69.1 (≥F2) 90.4 (≥F3) 96.9 (F4) | 90.9 (≥F2) 98.3 (≥F3) 88.0 (F4) | |
| Meng 2023 [ | Retrospective, single center | B-mode, SWE, clinical parameters | 618 patients | HR | SVM | Predicting the risk of fibrosis Progression NAFLD | LSM values | 0.95 | 86.2 | ||
| Li 2019 [ | Prospective, single center | B-mode, ORF and contrast-enhanced micro-flow (CEMF) | 144 patients | HR | Multiparametric model | Diagnosis of significant liver fibrosis | Liver surgery and biopsy | 0.78-0.85 | 87.5-93.8 | 69.2-76.9 | |
| Liu 2022 [ | Retrospective, single center | B-mode, liver stiffness values, and clinical parameters | 284 patients | DL | deep learning-based data integration network (DI-Net) | Diagnosis of significant liver fibrosis | Liver surgery and biopsy | 0.901 | 81.3 | 81.6 | 80.8 |
| Liu 2023 [ | Retrospective, single center | B-mode, Contrast-enhanced microflow (CEMF) cines and clinical parameters | 218 patients | DL | Data integration-based deep learning (DIDL) | Diagnosis of significant liver fibrosis | Liver surgery and biopsy | 0.901 | 81.3 | 81.6 | 80.9 |
| Gao 2021 [ | Retrospective, single center | B-mode, STQ, STE | 168 patients | DL | Multi-modal fusion network with AL (MMFN-AL) | Liver fibrosis classification | Liver biopsy | 0.891 | 70.59 | ||
| Xue 2020 [ | Retrospective, multicenter | B-mode, SWE | 466 patients | DL | Transfer learning of Inception-V3 | Liver fibrosis classification | Liver surgery | 0.93 (≥S2) 0.93 (≥S3) 0.95 (S4) | 90.0 (≥S2) 89.9 (≥S3) 90.1 (S4) | 87.8 (≥S2) 87.9 (≥S3) 94.3 (S4) | |
| Lu 2021 [ | Retrospective, multicenter | B-mode, SWE, clinical parameters | 807 patients | DL | Multichannel deep learning radiomics models (DLRE 2.0) | Diagnosis of significant liver fibrosis | Liver biopsy | 0.95 | 90.6 | 90.1 | |
| Chen 2024 [ | Retrospective, multicenter | B-mode, SWE | 5894 patients | DL | ResNet152 for liver stiffness prediction and sequential algorithm for liver fibrosis screening | Liver fibrosis screening | Liver biopsy | 85 | 54 | 94 | |
Table 3
Summary of published ultrasound radiomics studies for diagnosis of focal liver lesions"
| Reference | Study design | Imaging modality | No. of patients/ images | Radiomics technique | Detailed method | Task | Reference standard | Results | |||
| AUC | Accuracy | Sensitivity | Specificity | ||||||||
| HR, handcrafted radiomics; DL, deep learning; FLL, focal liver lesions; HCC, hepatocellular carcinoma; ICC, intrahepatic cholangiocarcinoma; FFS, focal fat sparing; FFI, focal fat infiltration; MLC, metastatic liver cancer; TIC, time-intensity curve; CEUS, contrast-enhanced ultrasound; CNN, convolutional neural networks; FNH, focal nodular hyperplasia; SWE, shear wave elastography; SWV, shear wave velocity | |||||||||||
| Hwang 2015 [ | Retrospective, NA | B-mode | 115 patients | HR | HR features with artificial neural network | FLL diagnosis (cysts, hemangiomas, malignancies) | NA | ||||
| Mao 2021 [ | Retrospective, single center | B-mode | 114 patients | HR | Different classifiers | Classification of primary and metastatic liver cancer | Pathology | 0.816 | 84.3 | 76.8 | 88 |
| Peng 2020 [ | Retrospective, single center | B-mode and clinical variables | 531 patients | HR | Different classifiers | Differentiation among HCC and ICC/cHCC-ICC | Pathology | 0.728-0.775 | |||
| Qin 2020 [ | Retrospective, single center | B-mode | 254 patients | HR | Different classifiers | Identification of primary tumorous sources of liver metastases | Pathology | 0.750-0.768 | 70.6; 79.2; 64.3 | 73.9; 70.0; 75.0 | 67.9; 85.7; 50.0 |
| Peng 2022 [ | Retrospective, single center | B-mode | 589 patients | HR | Different classifiers | Differentiating infected focal liver lesions from malignancy | Pathology or follow up/imaging | 0.745-0.836 | 66.7-78.4 | 65.4-87.7 | 45.2-67.7 |
| Schmauch 2019 [ | NA, | B-mode | 544 patients | DL | algorithm was trained using an attention with annotations | FLL diagnosis (benign and malignant) | NA | 0.812-0.922 | |||
| Xi 2021 [ | Retrospective, single center | B-mode | 596 patients | DL | Resnet | FLL diagnosis (benign and malignant) | MRI or histopathology | 0.85 | 80 | 91 | 62 |
| Tiyarattanachai 2021 [ | Retrospective, multicenter | B-mode | 22472 lesions | DL | RetinaNet for both detection and diagnosis | FLL detection and diagnosis (HCC, cyst, hemangioma, FFS and FFI) | Pathology and/or MRI/CT | 95.3 | 84.9 | 97.1 | |
| Chen 2024 [ | Retrospective, single center | B-mode | 465 patients | DL | Different DCNN | Differentiation among HCC, ICC, and cHCC-ICC | Pathology | 0.92 | 86 | 84.59 | 92.65 |
| Yang 2020 [ | Retrospective, multicenter | B-mode, clinical variables | 2143 patients | DL | Resnet 18 | FLL diagnosis (benign and malignant) | Pathology | 0.924 | 84.7 | 86.5 | 85.5 |
| Yang 2023 [ | Retrospective, multicenter | B-mode | 6784 patients | DL | Resnet 50 | Identification of hepatic echinococcosis | Pathological or clinical diagnosis | 0.913-0.982 (echinococcosis) 0.900-0.986 (alveolar echinococcosis) | 71.9-84; 90.7-94.6 | 92.1-100; 77.1-97.1 | 66.6-80.5; 92.4-94.2 |
| Du 2025 [ | Retrospective, multicenter | B-mode and clinical parameters | 1052 patients | HR | XGBoost | classification of ≤3 cm HCC | Pathology or CT/MRI with follow-up | 0.899 | 85.9 | 92.8 | 77.9 |
| Streba 2012 [ | Prospective, single center | CEUS | 112 patients | HR and DL | TIC features | FLL diagnosis (HCC, MLC, hepatic hemangiomas, local fatty changes) | Pathology or CT/MRI or follow up | 0.89 | 87.1 | 93.2 | 89.7 |
| Gatos 2015 [ | Retrospective, single center | CEUS | 52 patients | HR | TIC features | FLL detection and diagnosis (benign and malignant) | Pathology or CT/MRI | 0.89 | 90.3 | 93.1 | 86.9 |
| Kondo 2017 [ | Retrospective, single center | CEUS | 98 patients | HR | TIC features | FLL diagnosis (benign and malignant, differentiation of benign, HCC, or MLC) | Pathology or CT/MRI with follow-up | 91.8 | 94 | 87.1 | |
| Turco 2022 [ | Retrospective, single center | CEUS | 72 patients | HR | TIC features and textural features | FLL diagnosis (benign and malignant) | Pathology and/or MRI/CT | 0.84 | 84 | 76 | 92 |
| Guo 2018 [ | Retrospective, single center | CEUS | 93 patients | HR | Deep canonical correlation analysis (DCCA) and multiple kernel learning (MKL) | FLL diagnosis (benign and malignant) | Pathology or CT/MRI | 90.4 | 93.6 | 86.9 | |
| Li 2024 [ | Retrospective, single center | CEUS, clinical features | 159 patients | HR | Textual features from 6 CEUS images | Differentiation among HCC and ICC/cHCC-ICC in LR-M patients | Pathology | 0.912 | 85.4 | 95.2 | 77.8 |
| Wang 2024 [ | Prospective, multicenter | CEUS, clinical features | 116 patients | HR | HR features from one Kupffer phase image | Differentiating well-differentiated hepatocellular carcinoma (w-HCC) from atypical FLL | Pathology | 0.912 | 82.4 | 85 | |
| Pan 2019 [ | Retrospective, single center | CEUS | 242 tumors | DL | 3D-CNN | FLL diagnosis (HCC and FNH) | Pathology | 0.97 | 93.1 | 94.5 | 93.6 |
| Caleanu 2021 [ | Retrospective, single center | CEUS | 91 patients | DL | Different DCNN | FLL diagnosis (HCC, MLC, hemangiomas, FNH) | NA | 88 | |||
| Ding 2025 [ | Retrospective, multicenter | CEUS | 3725 lesions | DL | Module-Disease, Module-Biomarker and Module-Clinic were built and aggregated | FLL diagnosis (HCC, MLC, ICC, hepatic hemangioma, hepatic abscess and others) | Pathology for malignancy or MRI with follow-up for benign lesions | 0.86 | 85 | 97 | |
| Yao 2018 [ | Retrospective, single center | B-mode, SWE, SWV | 177 patients | HR | Sparse representation theory (SRT) | FLL diagnosis (benign and malignant) | Pathology | 0.94 | 88 | 91 | 86 |
| Hu 2024 [ | Retrospective, single center | B-mode, CEUS | 527 patients | HR | HR features extracted from both B-mode images and three-phase CEUS images | FLL diagnosis (benign and malignant) | Pathology for malignant lesions; CEUS and follow-up for benign lesions | 0.914 | 95.2 | 72.2 | |
| Su 2024 [ | Retrospective, single center | B-mode, CEUS, clinical features | 280 patients | HR | HR features from both B-mode and CEUS images | Differentiation of HCC and ICC | Pathology | 0.97 | 91 | 89 | 93 |
| Hu 2021 [ | Retrospective, single center | B-mode, CEUS | 574 patients | DL | ResNet on four-phase images | FLL diagnosis (benign and malignant) | Pathology for malignant lesions; CEUS and follow-up for benign lesions | 0.934 | 91 | 92.7 | 85.1 |
| Liu 2022 [ | Retrospective, multicenter | B-mode, CEUS, clinical features | 303 patients | DL | Four Stream (B-mode, CEUS and their optic flow) 3D convolutional neural network | FLL diagnosis (benign and malignant) | Pathology | 0.957 | 94 | 96.6 | 90.5 |
Table 4
Summary of published ultrasound radiomics studies for risk prediction of hepatocellular carcinoma"
| Reference | Study design | Imaging modality | No. of patients/images | Radiomics technique | Detailed method | Task | Reference standard | Results | |||
| AUC | Accuracy | Sensitivity | Specificity | ||||||||
| ORF, original radio frequency signal; HR, handcrafted radiomics; DL, deep learning; CEUS, contrast-enhanced ultrasound; MVI, microvascular invasion; CNN, convolutional neural networks; SWE, shear wave elastography | |||||||||||
| Dong 2019 [ | Prospective, single center | ORF | 42 patients | HR | Signal analysis and processing technology in feature extraction | MVI prediction | Pathology | 0.95 | 92.8 | 85.7 | 100 |
| Dong 2020 [ | Retrospective, single center | B-mode, clinical variables | 322 patients | HR | Gross- and peri-tumoral region HR | MVI prediction | Pathology | 0.744 | 63.4 | 89.2 | 48.4 |
| Hu 2019 [ | Retrospective, single center | B-mode, clinical variables | 482 patients | HR | Radiomics features combined with clinical variables | MVI prediction | Pathology | 0.731 | |||
| Dong 2022 [ | Prospective, single center | CEUS, clinical variables | 100 patients | HR | Radiomics features from peritumoral liver tissues of Kupffer phase | MVI prediction | Pathology | 0.804 | 75 | 87.5 | 69.1 |
| Zhang 2021 [ | Retrospective, single center | B-mode, CEUS, clinical variables | 313 patients | HR | HR features from different CEUS phase | MVI prediction | Pathology | 0.788 | 72.7 | 75.5 | 70.8 |
| Zhang 2022 [ | Retrospective, single center | CEUS, clinical variables | 436 patients | DL | GRU for temporal features and CNN for spatial features | MVI prediction | Pathology | 0.865 | 78.8 | 83.3 | 81 |
| Qin 2023 [ | Retrospective, single center | CEUS | 252 patients | DL | ResNet50+SE | MVI prediction | Pathology | 0.856 | 77.2 | 52.4 | 93.9 |
| Qin 2025 [ | Retrospective, single center | CEUS | 164 patients | DL | Transformer+Resnet | MVI prediction | Pathology | 0.859 | 71.4 | 86.2 | 80 |
| Wang 2025 [ | Retrospective, single center | CEUS | 318 patients | DL | Graph Convolutional Network | MVI prediction | Pathology | 0.928 | 89.3 | 85.3 | 91.7 |
| Zheng 2023 [ | Retrospective, single center | CEUS, MRI | 85 patients | HR | Comparison between CEUS and MRI radiomics model | MVI prediction | Pathology | 0.86 | 100 | 85.7 | |
| Zhang 2024 [ | Retrospective, multicenter | B-mode, CEUS, clinical variables | 576 patients | HR, DL | Comparison of DL and HR models with different conditions | MVI prediction | Pathology | 0.738 | 71 | 60.6 | 76.5 |
| Li 2025 [ | Experiments on rabbits | 3D ultrasound | 9 rabbits | HR | 3D US images are fused with whole-slide images to localize MVI regions | MVI prediction | Pathology | 0.91 | 86 | 76 | 92 |
| Ren 2021 [ | Retrospective, multicenter | B-mode, clinical variables | 193 patients | HR | combination of HR features and clinical variables | Differentiation prediction | Pathology | 0.849 | 81.8 | 75 | 85.7 |
| Qin 2023 [ | Retrospective, single center | CEUS | 272 patients | DL+HR | Combination of both DL and HR features | Differentiation prediction | Pathology | 0.932 | 91.5 | 93.8 | 90 |
| Li 2022 [ | Retrospective, single center | CEUS | 54 patients | HR | Radiomics features from Kupffer phase | Differentiation prediction | Pathology | 0.878-0.938 | 97.1 97.1 | 93.3 83.3 | 98.1 100 |
| Qian 2023 [ | Retrospective, single center | B-mode | 118 patients | HR | HR features from intratumoral and peritumoral regions | Ki-67 prediction | Pathology | 0.87 | 80.6 | 73.7 | 88.2 |
| Zhang 2024 [ | Retrospective, single center | B-mode, CEUS, clinical variables | 310 patients | HR | HR features from different CEUS phase | Ki-67 prediction | Pathology | 0.856 | 76.8 | 79.3 | 74.1 |
| Yao 2018 [ | Retrospective, single center | B-mode, SWE, SWV | 177 patients | HR | Sparse representation theory (SRT) | PD-1, Ki-67, MVI prediction | Pathology | 0.94-0.98 | 92; 93; 95 | 100; 95; 91 | 88; 89; 100 |
| Qian 2024 [ | Retrospective, single center | B-mode | 153 patients | HR | Different classifiers | Prediction of differentiation, CK-7, Ki-67 and p53 | Pathology | 0.762-0.922 | 82.1; 87.5; 75.0; 70.3 | 78.2; 100; 85.7 100 | |
| Bu 2025 [ | Retrospective, single center | B-mode, clinical parameters | 154 patients | HR | Different classifiers | Predicting TP53 mutation | Pathology | 0.846 | 82.3 | 80.6 | 83.9 |
| Liang 2025 [ | Retrospective, single center | B-mode, CEUS, clinical variables | 434 patients | HR | HR features from B-mode, arterial, portal venous, and delayed phases | CK-19 prediction | Pathology | 0.927 | 86.9 | 89.3 | 86.3 |
Table 5
Summary of published ultrasound radiomics studies for prognosis prediction of hepatocellular carcinoma"
| Reference | Study design | Imaging modality | No. of patients/images | Radiomics technique | Detailed method | Task | Reference standard | Results | ||||
| AUC | C-index | Accuracy | Sensitivity | Specificity | ||||||||
| ER, early recurrence; LR, late recurrence; RFS, recurrence free survival; MWA, microwave ablation; HR, handcrafted radiomics; DL, deep learning; CEUS, contrast-enhanced ultrasound; HCC, hepatocellular carcinoma; SR, surgical resection; RFA, radiofrequency ablation; PHLF, post-hepatectomy liver failure; ISGLS, international study group of liver surgery | ||||||||||||
| Huang 2021 [ | Retrospective, single center | B-mode, CEUS | 215 patients | HR | Combined features from both tumoral and peritumoral area in different CEUS phase | ER prediction after resection or ablation | follow up | 0.845 | 84.2 | 86.7 | 82.6 | |
| Cao 2024 [ | Retrospective, single center | B-mode, clinical variables | 127 patients | HR | Logistic regression | ER prediction following surgical resection | follow up | 0.925 | 77.8 | 100 | ||
| Wu 2022 [ | Retrospective, single center | B-mode | 513 patients | DL+HR | Comparison of DL and HR features | ER, LR, RFS after MWA and differentiation prediction | follow up | 0.695 (ER); 0.715 (LR); 0.721 (RFS) | ||||
| Zhang 2022 [ | Retrospective, single center | B-mode, CEUS, | 172 patients | DL+HR | Combination of DL and HR | ER prediction following surgical resection | follow up | 0.889 | 78.4 | 90 | 66.7 | |
| Huang 2022 [ | Retrospective, single center | B-mode, CEUS, clinical features | 414 patients | DL+HR | Combination of both HR and DL features | ER and survival prediction after resection | follow up | 0.57 (ER) | 0.759 (OS) | 59 (ER) | 62 (ER) | 56 (ER) |
| Ma 2021 [ | Retrospective, single center | B-mode, SWE, clinical features | 318 patients | DL+HR | Combining CEUS, US radiomics, and clinical factors | ER and LR prediction after ablation | follow up | 0.84 (ER) | 0.77 (LR) | 81 (ER) | 69 (ER) | 93 (ER) |
| Liu 2020 [ | Retrospective, single center | CEUS, clinical variables | 419 patients | DL | Cross Stratification Using R-RFA and R-SR in Swapped Groups | PFS prediction of early HCC after SR and RFA | follow up | 0.726 (RFA); 0.741 (SR) | ||||
| Zhong 2023 [ | Prospective, single center | SWE, clinical variables | 345 patients | HR | multi-scale radiomics model | PHLF prediction | ISGLS | 0.822 | 75 | 70.4 | 77.3 | |
| Xue 2024 [ | Retrospective, multi-center | B-mode, CEUS, clinical features | 532 patients | DL, HR | Resnet 50 trained with varying granularity with progressive training strategy | PHLF prediction | ISGLS | 0.860-1 | 88-90.5 | 75-100 | 87.5-100 | |
| Jiang 2025 [ | Retrospective, single center | SWE, clinical variables | 633 patients | HR | Different classifier | Postoperative complications and ER prediction after resection | Comprehensive Complication Index (CCI) and follow up | 0.832 (complications); 0.844 (ER) | 78; 66.3 | 78.6; 60.4 | 74.6: 98.5 | |
| Liu 2019 [ | Retrospective, single center | B-mode, CEUS | 130 patients | DL, HR | Comparison of R-DLCEUS, R-TIC, and R-Bmode models | Prediction of responses to TACE | mRECIST | 0.93 (R-DLCEUS) | 90 | 89.3 | 92.3 | |
| Jin 2021 [ | Retrospective, single center | B-mode, SWE, clinical features | 434 patients | DL | Combination of 2D-SWE and B-mode DL features, sex and age | HCC occurrence in chronic hepatitis B patients | follow up | 0.9 | 83.3 | 96.3 | ||
| [1] |
Asrani SK , Devarbhavi H , Eaton J , Kamath PS . Burden of liver diseases in the world. J Hepatol 2019; 70: 151-171.
doi: 10.1016/j.jhep.2018.09.014 |
| [2] | EASL Clinical Practice Guidelines on the management of hepatocellular carcinoma. J Hepatol 2025; 82:315-374. |
| [3] |
Lambin P , Leijenaar RTH , Deist TM , Peerlings J , de Jong EEC , van Timmeren J , et al . Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749-762.
doi: 10.1038/nrclinonc.2017.141 |
| [4] |
Zhang D , Zhang XY , Duan YY , Dietrich CF , Cui XW , Zhang CX . An overview of ultrasound-derived radiomics and deep learning in liver. Med Ultrason 2023; 25: 445-452.
doi: 10.11152/mu-4080 |
| [5] |
Quaia E , Calliada F , Bertolotto M , Rossi S , Garioni L , Rosa L , et al . Characterization of focal liver lesions with contrast-specific US modes and a sulfur hexafluoride-filled microbubble contrast agent: diagnostic performance and confidence. Radiology 2004; 232: 420-430.
doi: 10.1148/radiol.2322031401 |
| [6] |
Fang C , Sidhu PS . Ultrasound-based liver elastography: current results and future perspectives. Abdominal radiology (New York) 2020; 45: 3463-3472.
doi: 10.1007/s00261-020-02717-x |
| [7] | Zhang Y , Cui J , Wan W , Liu J . Multimodal imaging under artificial intelligence algorithm for the diagnosis of liver cancer and its relationship with expressions of EZH2 and p57. Comput Intell Neurosci 2022; 2022: 4081654 |
| [8] |
Xue LY , Jiang ZY , Fu TT , Wang QM , Zhu YL , Dai M , et al . Transfer learning radiomics based on multimodal ultrasound imaging for staging liver fibrosis. Eur Radiol 2020; 30: 2973-2983.
doi: 10.1007/s00330-019-06595-w |
| [9] | Wei X , Wang Y , Wang L , Gao M , He Q , Zhang Y , et al. Simultaneous grading diagnosis of liver fibrosis, inflammation, and steatosis using multimodal quantitative ultrasound and artificial intelligence framework. Med Biol Eng Comput 2024. |
| [10] |
Lambin P , Rios-Velazquez E , Leijenaar R , Carvalho S , Van Stiphout RG , Granton P , et al . Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441-446.
doi: 10.1016/j.ejca.2011.11.036 |
| [11] | Nohara Y , Matsumoto K , Soejima H , Nakashima NJCM , Biomedicine Pi . Explanation of machine learning models using shapley additive explanation and application for real data in hospital Comput Methods Programs Biomed 2022; 214:106584. |
| [12] | Traverso A , Wee L , Dekker A , Gillies RJIJoROBP . Repeatability and reproducibility of radiomic features: a systematic review Int J Radiat Oncol Biol Phys 2018; 102:1143-1158. |
| [13] |
Lecun Y , Bottou L , Bengio Y , Haffner P . Gradient-based learning applied to document recognition. Proceedings of the IEEE 1998; 86: 2278-2324.
doi: 10.1109/5.726791 |
| [14] |
Hochreiter S , Schmidhuber J . Long Short-Term Memory. Neural Computation 1997; 9: 1735-1780.
doi: 10.1162/neco.1997.9.8.1735 |
| [15] |
Wu Z , Pan S , Chen F , Long G , Zhang C , Yu PS . A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Syst 2021; 32: 4-24.
doi: 10.1109/TNNLS.2020.2978386 |
| [16] | Vaswani A , Shazeer N , Parmar N , Uszkoreit J , Jones L , Gomez AN , et al. Attention is all you need. 2017;30. |
| [17] | Dosovitskiy A , Beyer L , Kolesnikov A , Weissenborn D , Zhai X , Unterthiner T , et al. An image is worth 16x16 words: Transformers for image recognition at scale. 2020. |
| [18] | Salahuddin Z , Woodruff HC , Chatterjee A , Lambin PJCib , medicine. Transparency of deep neural networks for medical image analysis: A review of interpretability methods Comput Biol Med 2022; 140:105111. |
| [19] |
Li MD , Cheng MQ , Chen LD , Hu HT , Zhang JC , Ruan SM , et al . Reproducibility of radiomics features from ultrasound images: influence of image acquisition and processing. Eur Radiol 2022; 32: 5843-5851.
doi: 10.1007/s00330-022-08662-1 |
| [20] |
Salem N , Malik H , Shams A . Medical image enhancement based on histogram algorithms. Procedia Computer Science 2019; 163: 300-311.
doi: 10.1016/j.procs.2019.12.112 |
| [21] |
Rhyou SY , Yoo JC . Cascaded deep learning neural network for automated liver steatosis diagnosis using ultrasound images. Sensors (Basel, Switzerland) 2021; 21: 5304.
doi: 10.3390/s21165304 |
| [22] |
Mali SA , Ibrahim A , Woodruff HC , Andrearczyk V , Müller H , Primakov S , et al . Making radiomics more reproducible across scanner and imaging protocol variations: a review of harmonization methods. J Pers Med 2021; 11: 842.
doi: 10.3390/jpm11090842 |
| [23] | Zhao Z , Qin Y , Shao K , Liu Y , Zhang Y , Li H , et al. Radiomics harmonization in ultrasound images for cervical cancer lymph node metastasis prediction using Cycle-GAN. Technol Cancer Res Treat 2024; 23:15330338241302237. |
| [24] |
Johnson WE , Li C , Rabinovic A . Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007; 8: 118-127.
doi: 10.1093/biostatistics/kxj037 |
| [25] |
Duarte-Salazar CA , Castro-Ospina AE , Becerra MA , Delgado-Trejos E . Speckle noise reduction in ultrasound images for improving the metrological evaluation of biomedical applications: an overview. IEEE Access 2020; 8: 15983-15999.
doi: 10.1109/ACCESS.2020.2967178 |
| [26] |
Dietrich CF , Nolsøe CP , Barr RG , Berzigotti A , Burns PN , Cantisani V , et al . Guidelines and good clinical practice recommendations for contrast-enhanced ultrasound (CEUS) in the liver-update 2020 WFUMB in cooperation with EFSUMB, AFSUMB, AIUM, and FLAUS. Ultrasound Med Biol 2020; 46: 2579-2604.
doi: 10.1016/j.ultrasmedbio.2020.04.030 |
| [27] |
Tian H , Cai W , Ding W , Liang P , Yu J , Huang Q . Long-term liver lesion tracking in contrast-enhanced ultrasound videos via a siamese network with temporal motion attention. Front Physiol 2023; 14: 1180713.
doi: 10.3389/fphys.2023.1180713 |
| [28] |
Li MD , Hu HT , Ruan SM , Cheng MQ , Chen LD , Huang ZR , et al . ADMNet: adaptive-weighting dual mapping for online tracking with respiratory motion estimation in contrast-enhanced ultrasound. IEEE Trans Image Process 2024; 33: 58-68.
doi: 10.1109/TIP.2023.3333195 |
| [29] |
Duan Y , Shi S , Long H , Zhong X , Tan Y , Liu G , et al . A bi-modal temporal segmentation network for automated segmentation of focal liver lesions in dynamic contrast-enhanced ultrasound. Ultrasound Med Biol 2025; 51: 759-767.
doi: 10.1016/j.ultrasmedbio.2024.12.014 |
| [30] | Dietrich CF , Bamber J , Berzigotti A , Bota S , Cantisani V , Castera L , et al . EFSUMB guidelines and recommendations on the clinical use of liver ultrasound elastography, update 2017 (long version). Ultraschall Med 2017; 38: e16-e47 |
| [31] |
Ferraioli G , Filice C , Castera L , Choi BI , Sporea I , Wilson SR , et al . WFUMB guidelines and recommendations for clinical use of ultrasound elastography: Part 3: liver. Ultrasound Med Biol 2015; 41: 1161-1179.
doi: 10.1016/j.ultrasmedbio.2015.03.007 |
| [32] |
Wang H , Zheng P , Wang X , Sang L . Effect of Q-Box size on liver stiffness measurement by two-dimensional shear wave elastography. J Clin Ultrasound 2021; 49: 978-983.
doi: 10.1002/jcu.23075 |
| [33] |
Wang K , Lu X , Zhou H , Gao Y , Zheng J , Tong M , et al . Deep learning radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut 2019; 68: 729-741.
doi: 10.1136/gutjnl-2018-316204 |
| [34] |
Liu Z , Li W , Zhu Z , Wen H , Li MD , Hou C , et al . A deep learning model with data integration of ultrasound contrast-enhanced micro-flow cines, B-mode images, and clinical parameters for diagnosing significant liver fibrosis in patients with chronic hepatitis B. Eur Radiol 2023; 33: 5871-5881.
doi: 10.1007/s00330-023-09436-z |
| [35] |
Li W , Huang Y , Zhuang BW , Liu GJ , Hu HT , Li X , et al . Multiparametric ultrasomics of significant liver fibrosis: A machine learning-based analysis. Eur Radiol 2019; 29: 1496-1506.
doi: 10.1007/s00330-018-5680-z |
| [36] |
Stahlschmidt SR , Ulfenborg B , Synnergren J . Multimodal deep learning for biomedical data fusion: a review. Brief Bioinform 2022; 23: bbab569.
doi: 10.1093/bib/bbab569 |
| [37] |
Younossi ZM , Golabi P , Paik JM , Henry A , Van Dongen C , Henry L . The global epidemiology of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH): a systematic review. Hepatology 2023; 77: 1335-1347.
doi: 10.1097/HEP.0000000000000004 |
| [38] | Wu CH , Hung CL , Lee TY , Wu CY , Chu WCC , editors. Fatty liver diagnosis using deep learning in ultrasound image. 2022 IEEE International Conference on Digital Health (ICDH) 2022 10-16 July 2022. |
| [39] |
Cao W , An X , Cong L , Lyu C , Zhou Q , Guo R . Application of deep learning in quantitative analysis of 2-dimensional ultrasound imaging of nonalcoholic fatty liver disease. J Ultrasound Med 2020; 39: 51-59.
doi: 10.1002/jum.15070 |
| [40] |
Byra M , Styczynski G , Szmigielski C , Kalinowski P , Michałowski Ł , Paluszkiewicz R , et al . Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg 2018; 13: 1895-1903.
doi: 10.1007/s11548-018-1843-2 |
| [41] | Reddy DS , Bharath R , Rajalakshmi P , editors. A novel computer-aided diagnosis framework using deep learning for classification of fatty liver disease in ultrasound imaging. 2018 IEEE 20th International Conference on e-Health Networking, Applications and Services (Healthcom) 2018 17-20 Sept. 2018. |
| [42] |
Kim T , Lee DH , Park EK , Choi S . Deep learning techniques for fatty liver using multi-view ultrasound images scanned by different scanners: development and validation study. JMIR Med Inform 2021; 9: e30066.
doi: 10.2196/30066 |
| [43] | Han A , Byra M , Heba E , Andre MP , Erdman JW , Jr R , et al . Noninvasive diagnosis of nonalcoholic fatty liver disease and quantification of liver fat with radiofrequency ultrasound data using one-dimensional convolutional neural networks. Radiology 2020; 295: 342-350 |
| [44] |
Chou TH , Yeh HJ , Chang CC , Tang JH , Kao WY , Su IC , et al . Deep learning for abdominal ultrasound: a computer-aided diagnostic system for the severity of fatty liver. J Chin Med Assoc 2021; 84: 842-850.
doi: 10.1097/JCMA.0000000000000585 |
| [45] |
Tahmasebi A , Wang S , Wessner CE , Vu T , Liu JB , Forsberg F , et al . Ultrasound-based machine learning approach for detection of nonalcoholic fatty liver disease. J Ultrasound Med 2023; 42: 1747-1756.
doi: 10.1002/jum.16194 |
| [46] |
Yang Y , Liu J , Sun C , Shi Y , Hsing JC , Kamya A , et al . Nonalcoholic fatty liver disease (NAFLD) detection and deep learning in a Chinese community-based population. Eur Radiol 2023; 33: 5894-5906.
doi: 10.1007/s00330-023-09515-1 |
| [47] |
Liu Y , Yu W , Wang P , Huang Y , Li J , Li P . Deep learning with ultrasound images enhance the diagnosis of nonalcoholic fatty liver. Ultrasound Med Biol 2024; 50: 1724-1730.
doi: 10.1016/j.ultrasmedbio.2024.07.014 |
| [48] | Meng D , Zhang L , Cao G , Cao W , Zhang G , Hu B . Liver fibrosis classification based on transfer learning and fcnet for ultrasound images. IEEE Access 2017; 5: 5804-5810 |
| [49] |
Lee JH , Joo I , Kang TW , Paik YH , Sinn DH , Ha SY , et al . Deep learning with ultrasonography: automated classification of liver fibrosis using a deep convolutional neural network. Eur Radiol 2020; 30: 1264-1273.
doi: 10.1007/s00330-019-06407-1 |
| [50] |
Feng X , Chen X , Dong C , Liu Y , Liu Z , Ding R , et al . Multi-scale information with attention integration for classification of liver fibrosis in B-mode US image. Comput Methods Programs Biomed 2022; 215: 106598.
doi: 10.1016/j.cmpb.2021.106598 |
| [51] |
Ruan D , Shi Y , Jin L , Yang Q , Yu W , Ren H , et al . An ultrasound image-based deep multi-scale texture network for liver fibrosis grading in patients with chronic HBV infection. Liver Int 2021; 41: 2440-2454.
doi: 10.1111/liv.14999 |
| [52] |
Duan YY , Qin J , Qiu WQ , Li SY , Li C , Liu AS , et al . Performance of a generative adversarial network using ultrasound images to stage liver fibrosis and predict cirrhosis based on a deep-learning radiomics nomogram. Clin Radiol 2022; 77: e723-e731.
doi: 10.1016/j.crad.2022.06.003 |
| [53] |
Joo Y , Park HC , Lee OJ , Yoon C , Choi MH , Choi C . Classification of liver fibrosis from heterogeneous ultrasound image. IEEE Access 2023; 11: 9920-9930.
doi: 10.1109/ACCESS.2023.3240216 |
| [54] |
Park HC , Joo Y , Lee OJ , Lee K , Song TK , Choi C , et al . Automated classification of liver fibrosis stages using ultrasound imaging. BMC Med Imaging 2024; 24: 36.
doi: 10.1186/s12880-024-01209-4 |
| [55] |
Ai H , Huang Y , Tai DI , Tsui PH , Zhou Z . Ultrasonic assessment of liver fibrosis using one-dimensional convolutional neural networks based on frequency spectra of radiofrequency signals with deep learning segmentation of liver regions in B-mode images: a feasibility study. Sensors (Basel, Switzerland) 2024; 24: 5513.
doi: 10.3390/s24175513 |
| [56] |
Zhang L , Tan Z , Li C , Mou L , Shi YL , Zhu XX , et al . A deep learning model based on high-frequency ultrasound images for classification of different stages of liver fibrosis. Liver Int 2025; 45: e70148.
doi: 10.1111/liv.70148 |
| [57] |
Chen Y , Luo Y , Huang W , Hu D , Zheng RQ , Cong SZ , et al . Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med 2017; 89: 18-23.
doi: 10.1016/j.compbiomed.2017.07.012 |
| [58] |
Gatos I , Tsantis S , Spiliopoulos S , Karnabatidis D , Theotokas I , Zoumpoulis P , et al . A new computer aided diagnosis system for evaluation of chronic liver disease with ultrasound shear wave elastography imaging. Medical physics 2016; 43: 1428-1436.
doi: 10.1118/1.4942383 |
| [59] |
Gatos I , Tsantis S , Spiliopoulos S , Karnabatidis D , Theotokas I , Zoumpoulis P , et al . A machine-learning algorithm toward color analysis for chronic liver disease classification, employing ultrasound shear wave elastography. Ultrasound Med Biol 2017; 43: 1797-1810.
doi: 10.1016/j.ultrasmedbio.2017.05.002 |
| [60] |
Gatos I , Tsantis S , Spiliopoulos S , Karnabatidis D , Theotokas I , Zoumpoulis P , et al . Temporal stability assessment in shear wave elasticity images validated by deep learning neural network for chronic liver disease fibrosis stage assessment. Medical physics 2019; 46: 2298-2309.
doi: 10.1002/mp.13521 |
| [61] |
Kagadis GC , Drazinos P , Gatos I , Tsantis S , Papadimitroulas P , Spiliopoulos S , et al . Deep learning networks on chronic liver disease assessment with fine-tuning of shear wave elastography image sequences. Phys Med Biol 2020; 65: 215027.
doi: 10.1088/1361-6560/abae06 |
| [62] |
Meng F , Wu Q , Zhang W , Hou S . Application of interpretable machine learning models based on ultrasonic radiomics for predicting the risk of fibrosis progression in diabetic patients with nonalcoholic fatty liver disease. Diabetes Metab Syndr Obes 2023; 16: 3901-3913.
doi: 10.2147/DMSO.S439127 |
| [63] |
Liu Z , Wen H , Zhu Z , Li Q , Liu L , Li T , et al . Diagnosis of significant liver fibrosis in patients with chronic hepatitis B using a deep learning-based data integration network. Hepatol Int 2022; 16: 526-536.
doi: 10.1007/s12072-021-10294-4 |
| [64] | Gao L , Zhou R , Dong C , Feng C , Li Z , Wan X , et al., editors. Multi-modal active learning for automatic liver fibrosis diagnosis based on ultrasound shear wave elastography. 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) 2021 13-16 April 2021. |
| [65] |
Lu X , Zhou H , Wang K , Jin J , Meng F , Mu X , et al . Comparing radiomics models with different inputs for accurate diagnosis of significant fibrosis in chronic liver disease. Eur Radiol 2021; 31: 8743-8754.
doi: 10.1007/s00330-021-07934-6 |
| [66] |
Chen LD , Huang ZR , Yang H , Cheng MQ , Hu HT , Lu XZ , et al . US-based sequential algorithm integrating an ai model for advanced liver fibrosis screening. Radiology 2024; 311: e231461.
doi: 10.1148/radiol.231461 |
| [67] | Hwang YN , Lee JH , Kim GY , Jiang YY , Kim SM . Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng 2015;26 Suppl 1:S1599-611. |
| [68] |
Mao B , Ma J , Duan S , Xia Y , Tao Y , Zhang L . Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur Radiol 2021; 31: 4576-4586.
doi: 10.1007/s00330-020-07562-6 |
| [69] |
Peng Y , Lin P , Wu L , Wan D , Zhao Y , Liang L , et al . Ultrasound-based radiomics analysis for preoperatively predicting different histopathological subtypes of primary liver cancer. Frontiers in oncology 2020; 10: 1646.
doi: 10.3389/fonc.2020.01646 |
| [70] |
Qin H , Wu YQ , Lin P , Gao RZ , Li X , Wang XR , et al . Ultrasound image-based radiomics: an innovative method to identify primary tumorous sources of liver metastases. J Ultrasound Med 2021; 40: 1229-1244.
doi: 10.1002/jum.15506 |
| [71] |
Peng JB , Peng YT , Lin P , Wan D , Qin H , Li X , et al . Differentiating infected focal liver lesions from malignant mimickers: value of ultrasound-based radiomics. Clin Radiol 2022; 77: 104-113.
doi: 10.1016/j.crad.2021.10.009 |
| [72] |
Xi IL , Wu J , Guan J , Zhang PJ , Horii SC , Soulen MC , et al . Deep learning for differentiation of benign and malignant solid liver lesions on ultrasonography. Abdom Radiol (NY) 2021; 46: 534-543.
doi: 10.1007/s00261-020-02564-w |
| [73] |
Schmauch B , Herent P , Jehanno P , Dehaene O , Saillard C , Aubé C , et al . Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging 2019; 100: 227-233.
doi: 10.1016/j.diii.2019.02.009 |
| [74] | Chen J , Zhang W , Bao J , Wang K , Zhao Q , Zhu Y , et al . Implications of ultrasound-based deep learning model for preoperatively differentiating combined hepatocellular-cholangiocarcinoma from hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Abdom Radiol (NY) 2024; 49: 93-102 |
| [75] |
Tiyarattanachai T , Apiparakoon T , Marukatat S , Sukcharoen S , Geratikornsupuk N , Anukulkarnkusol N , et al . Development and validation of artificial intelligence to detect and diagnose liver lesions from ultrasound images. PLoS One 2021; 16: e0252882.
doi: 10.1371/journal.pone.0252882 |
| [76] |
Yang Q , Wei J , Hao X , Kong D , Yu X , Jiang T , et al . Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: A multicentre study. EBioMedicine 2020; 56: 102777.
doi: 10.1016/j.ebiom.2020.102777 |
| [77] |
Yang Y , Cairang Y , Jiang T , Zhou J , Zhang L , Qi B , et al . Ultrasound identification of hepatic echinococcosis using a deep convolutional neural network model in China: a retrospective, large-scale, multicentre, diagnostic accuracy study. Lancet Digit Health 2023; 5: e503-e514.
doi: 10.1016/S2589-7500(23)00091-2 |
| [78] |
Du Z , Fan F , Ma J , Liu J , Yan X , Chen X , et al . Development and validation of an ultrasound-based interpretable machine learning model for the classification of ≤ 3 cm hepatocellular carcinoma: a multicentre retrospective diagnostic study. EClinicalMedicine 2025; 81: 103098.
doi: 10.1016/j.eclinm.2025.103098 |
| [79] |
Streba CT , Ionescu M , Gheonea DI , Sandulescu L , Ciurea T , Saftoiu A , et al . Contrast-enhanced ultrasonography parameters in neural network diagnosis of liver tumors. World J Gastroenterol 2012; 18: 4427-4434.
doi: 10.3748/wjg.v18.i32.4427 |
| [80] |
Gatos I , Tsantis S , Spiliopoulos S , Skouroliakou A , Theotokas I , Zoumpoulis P , et al . A new automated quantification algorithm for the detection and evaluation of focal liver lesions with contrast-enhanced ultrasound. Medical physics 2015; 42: 3948-3959.
doi: 10.1118/1.4921753 |
| [81] |
Kondo S , Takagi K , Nishida M , Iwai T , Kudo Y , Ogawa K , et al . Computer-aided diagnosis of focal liver lesions using contrast-enhanced ultrasonography with perflubutane microbubbles. IEEE Trans Med Imaging 2017; 36: 1427-1437.
doi: 10.1109/TMI.2017.2659734 |
| [82] |
Turco S , Tiyarattanachai T , Ebrahimkheil K , Eisenbrey J , Kamaya A , Mischi M , et al . Interpretable machine learning for characterization of focal liver lesions by contrast-enhanced ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control 2022; 69: 1670-1681.
doi: 10.1109/TUFFC.2022.3161719 |
| [83] |
Guo LH , Wang D , Qian YY , Zheng X , Zhao CK , Li XL , et al . A two-stage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images. Clin Hemorheol Microcirc 2018; 69: 343-354.
doi: 10.3233/CH-170275 |
| [84] |
Li L , Liang X , Yu Y , Mao R , Han J , Peng C , et al . Radiomics-based machine learning classification strategy for characterization of hepatocellular carcinoma on contrast-enhanced ultrasound in high-risk patients with li-rads category m nodules. Indian J Radiol Imaging 2024; 34: 405-415.
doi: 10.1055/s-0043-1777993 |
| [85] |
Wang Z , Yao J , Jing X , Li K , Lu S , Yang H , et al . A combined model based on radiomics features of Sonazoid contrast-enhanced ultrasound in the Kupffer phase for the diagnosis of well-differentiated hepatocellular carcinoma and atypical focal liver lesions: a prospective, multicenter study. Abdom Radiol (NY) 2024; 49: 3427-3437.
doi: 10.1007/s00261-024-04253-4 |
| [86] | Pan F , Huang Q , Li X , editors. Classification of liver tumors with CEUS based on 3D-CNN. 2019 IEEE 4th International Conference on Advanced Robotics and Mechatronics (ICARM) 2019 3-5 July 2019. |
| [87] |
Căleanu CD , Sîrbu CL , Simion G . Deep neural architectures for contrast enhanced ultrasound (ceus) focal liver lesions automated diagnosis. Sensors (Basel, Switzerland) 2021; 21: 4126.
doi: 10.3390/s21124126 |
| [88] |
Ding W , Meng Y , Ma J , Pang C , Wu J , Tian J , et al . Contrast-enhanced ultrasound-based AI model for multi-classification of focal liver lesions. Journal of Hepatology 2025; 83: 426-439.
doi: 10.1016/j.jhep.2025.01.011 |
| [89] |
Yao Z , Dong Y , Wu G , Zhang Q , Yang D , Yu JH , et al . Preoperative diagnosis and prediction of hepatocellular carcinoma: Radiomics analysis based on multi-modal ultrasound images. BMC Cancer 2018; 18: 1089.
doi: 10.1186/s12885-018-5003-4 |
| [90] |
Hu HT , Li MD , Zhang JC , Ruan SM , Wu SS , Lin XX , et al . Ultrasomics differentiation of malignant and benign focal liver lesions based on contrast-enhanced ultrasound. BMC medical imaging 2024; 24: 242.
doi: 10.1186/s12880-024-01426-x |
| [91] |
Su LY , Xu M , Chen YL , Lin MX , Xie XY . Ultrasomics in liver cancer: developing a radiomics model for differentiating intrahepatic cholangiocarcinoma from hepatocellular carcinoma using contrast-enhanced ultrasound. World J Radiol 2024; 16: 247-255.
doi: 10.4329/wjr.v16.i7.247 |
| [92] |
Hu HT , Wang W , Chen LD , Ruan SM , Chen SL , Li X , et al . Artificial intelligence assists identifying malignant versus benign liver lesions using contrast-enhanced ultrasound. J Gastroenterol Hepatol 2021; 36: 2875-2883.
doi: 10.1111/jgh.15522 |
| [93] |
Liu L , Tang C , Li L , Chen P , Tan Y , Hu X , et al . Deep learning radiomics for focal liver lesions diagnosis on long-range contrast-enhanced ultrasound and clinical factors. Quant Imaging Med Surg 2022; 12: 3213-3226.
doi: 10.21037/qims-21-1004 |
| [94] |
Dong Y , Wang QM , Li Q , Li LY , Zhang Q , Yao Z , et al . Preoperative prediction of microvascular invasion of hepatocellular carcinoma: radiomics algorithm based on ultrasound original radio frequency signals. Frontiers in oncology 2019; 9: 1203.
doi: 10.3389/fonc.2019.01203 |
| [95] |
Dong Y , Zhou L , Xia W , Zhao XY , Zhang Q , Jian JM , et al . Preoperative prediction of microvascular invasion in hepatocellular carcinoma: initial application of a radiomic algorithm based on grayscale ultrasound images. Frontiers in oncology 2020; 10: 353.
doi: 10.3389/fonc.2020.00353 |
| [96] |
Hu HT , Wang Z , Huang XW , Chen SL , Zheng X , Ruan SM , et al . Ultrasound-based radiomics score: a potential biomarker for the prediction of microvascular invasion in hepatocellular carcinoma. Eur Radiol 2019; 29: 2890-2901.
doi: 10.1007/s00330-018-5797-0 |
| [97] |
Dong Y , Zuo D , Qiu YJ , Cao JY , Wang HZ , Yu LY , et al . Preoperative prediction of microvascular invasion (MVI) in hepatocellular carcinoma based on kupffer phase radiomics features of sonazoid contrast-enhanced ultrasound (SCEUS): A prospective study. Clinical hemorheology and microcirculation 2022; 81: 97-107.
doi: 10.3233/CH-211363 |
| [98] |
Zhang D , Wei Q , Wu GG , Zhang XY , Lu WW , Lv WZ , et al . Preoperative prediction of microvascular invasion in patients with hepatocellular carcinoma based on radiomics nomogram using contrast-enhanced ultrasound. Frontiers in oncology 2021; 11: 709339.
doi: 10.3389/fonc.2021.709339 |
| [99] |
Zhang Y , Wei Q , Huang Y , Yao Z , Yan C , Zou X , et al . Deep learning of liver contrast-enhanced ultrasound to predict microvascular invasion and prognosis in hepatocellular carcinoma. Frontiers in oncology 2022; 12: 878061.
doi: 10.3389/fonc.2022.878061 |
| [100] | Qin X , Zhu J , Tu Z , Ma Q , Tang J , Zhang C . Contrast-enhanced ultrasound with deep learning with attention mechanisms for predicting microvascular invasion in single hepatocellular carcinoma. Academic radiology 2023;30 Suppl 1:S73-S80. |
| [101] |
Qin Q , Pang J , Li J , Gao R , Wen R , Wu Y , et al . Transformer model based on Sonazoid contrast-enhanced ultrasound for microvascular invasion prediction in hepatocellular carcinoma. Medical physics 2025; 52: e17895.
doi: 10.1002/mp.17895 |
| [102] |
Wang Y , Xie W , Li C , Xu Q , Du Z , Zhong Z , et al . Automated microvascular invasion prediction of hepatocellular carcinoma via deep relation reasoning from dynamic contrast-enhanced ultrasound. Computerized medical imaging and graphics: the official journal of the Computerized Medical Imaging Society 2025; 124: 102606.
doi: 10.1016/j.compmedimag.2025.102606 |
| [103] |
Zheng R , Zhang X , Liu B , Zhang Y , Shen H , Xie X , et al . Comparison of non-radiomics imaging features and radiomics models based on contrast-enhanced ultrasound and Gd-EOB-DTPA-enhanced MRI for predicting microvascular invasion in hepatocellular carcinoma within 5 cm. Eur Radiol 2023; 33: 6462-6472.
doi: 10.1007/s00330-023-09789-5 |
| [104] | Zhang W , Guo Q , Zhu Y , Wang M , Zhang T , Cheng G , et al . Cross-institutional evaluation of deep learning and radiomics models in predicting microvascular invasion in hepatocellular carcinoma: validity, robustness, and ultrasound modality efficacy comparison. Cancer imaging : the official publication of the International Cancer Imaging Society 2024; 24: 142 |
| [105] |
Li L , Wang S , Chen J , Wu C , Chen Z , Ye F , et al . Radiomics diagnosis of microvascular invasion in hepatocellular carcinoma using 3D ultrasound and whole-slide image fusion. Small methods 2025; 9: e2401617.
doi: 10.1002/smtd.202401617 |
| [106] |
Ren S , Qi Q , Liu S , Duan S , Mao B , Chang Z , et al . Preoperative prediction of pathological grading of hepatocellular carcinoma using machine learning-based ultrasomics: A multicenter study. Eur J Radiol 2021; 143: 109891.
doi: 10.1016/j.ejrad.2021.109891 |
| [107] |
Qin X , Hu X , Xiao W , Zhu C , Ma Q , Zhang C . Preoperative evaluation of hepatocellular carcinoma differentiation using contrast-enhanced ultrasound-based deep-learning radiomics model. Journal of hepatocellular carcinoma 2023; 10: 157-168.
doi: 10.2147/JHC.S400166 |
| [108] | Li C , Xu J , Liu Y , Wu M , Dai W , Song J , et al . Kupffer phase radiomics signature in sonazoid-enhanced ultrasound is an independent and effective predictor of the pathologic grade of hepatocellular carcinoma. Journal of oncology 2022; 2022: 6123242 |
| [109] |
Qian H , Shen Z , Zhou D , Huang Y . Intratumoral and peritumoral radiomics model based on abdominal ultrasound for predicting Ki-67 expression in patients with hepatocellular cancer. Frontiers in oncology 2023; 13: 1209111.
doi: 10.3389/fonc.2023.1209111 |
| [110] |
Zhang D , Zhang XY , Lu WW , Liao JT , Zhang CX , Tang Q , et al . Predicting Ki-67 expression in hepatocellular carcinoma: nomogram based on clinical factors and contrast-enhanced ultrasound radiomics signatures. Abdominal radiology (New York) 2024; 49: 1419-1431.
doi: 10.1007/s00261-024-04191-1 |
| [111] |
Qian H , Huang Y , Xu L , Fu H , Lu B . Role of peritumoral tissue analysis in predicting characteristics of hepatocellular carcinoma using ultrasound-based radiomics. Sci Rep 2024; 14: 11538.
doi: 10.1038/s41598-024-62457-6 |
| [112] | Bu D , Duan S , Ren S , Ma Y , Liu Y , Li Y , et al . Machine learning-based ultrasound radiomics for predicting TP53 mutation status in hepatocellular carcinoma. Front Med (Lausanne) 2025; 12: 1565618 |
| [113] |
Liang L , Pang JS , Gao RZ , Que Q , Wu YQ , Peng JB , et al . Development and validation of a combined radiomic and clinical model based on contrast-enhanced ultrasound for preoperative prediction of CK19-positive hepatocellular carcinoma. Abdominal radiology (New York) 2025; 50: 3516-3529.
doi: 10.1007/s00261-025-04799-x |
| [114] |
Huang H , Ruan SM , Xian MF , Li MD , Cheng MQ , Li W , et al . Contrast-enhanced ultrasound-based ultrasomics score: a potential biomarker for predicting early recurrence of hepatocellular carcinoma after resection or ablation. The British journal of radiology 2022; 95: 20210748.
doi: 10.1259/bjr.20210748 |
| [115] |
Cao K , Wang X , Xu C , Wu L , Li L , Yuan Y , et al . Ultrasound-based radiomics analysis for assessing risk factors associated with early recurrence following surgical resection of hepatocellular carcinoma. Ultrasound Med Biol 2024; 50: 1964-1972.
doi: 10.1016/j.ultrasmedbio.2024.09.002 |
| [116] |
Wu JP , Ding WZ , Wang YL , Liu S , Zhang XQ , Yang Q , et al . Radiomics analysis of ultrasound to predict recurrence of hepatocellular carcinoma after microwave ablation. International journal of hyperthermia: the official journal of European Society for Hyperthermic Oncology, North American Hyperthermia Group 2022; 39: 595-604.
doi: 10.1080/02656736.2022.2062463 |
| [117] |
Zhang H , Huo F . Prediction of early recurrence of HCC after hepatectomy by contrast-enhanced ultrasound-based deep learning radiomics. Frontiers in oncology 2022; 12: 930458.
doi: 10.3389/fonc.2022.930458 |
| [118] |
Huang Z , Shu Z , Zhu RH , Xin JY , Wu LL , Wang HZ , et al . Deep learning-based radiomics based on contrast-enhanced ultrasound predicts early recurrence and survival outcome in hepatocellular carcinoma. World journal of gastrointestinal oncology 2022; 14: 2380-2392.
doi: 10.4251/wjgo.v14.i12.2380 |
| [119] |
Ma QP , He XL , Li K , Wang JF , Zeng QJ , Xu EJ , et al . Dynamic contrast-enhanced ultrasound radiomics for hepatocellular carcinoma recurrence prediction after thermal ablation. Molecular imaging and biology 2021; 23: 572-585.
doi: 10.1007/s11307-021-01578-0 |
| [120] |
Liu F , Liu D , Wang K , Xie X , Su L , Kuang M , et al . Deep learning radiomics based on contrast-enhanced ultrasound might optimize curative treatments for very-early or early-stage hepatocellular carcinoma patients. Liver Cancer 2020; 9: 397-413.
doi: 10.1159/000505694 |
| [121] |
Zhong X , Salahuddin Z , Chen Y , Woodruff HC , Long H , Peng J , et al . An interpretable radiomics model based on two-dimensional shear wave elastography for predicting symptomatic post-hepatectomy liver failure in patients with hepatocellular carcinoma. Cancers 2023; 15: 5303.
doi: 10.3390/cancers15215303 |
| [122] |
Xue L , Zhu J , Fang Y , Xie X , Cheng G , Zhang Y , et al . Preoperative ultrasound radomics to predict posthepatectomy liver failure in patients with hepatocellular carcinoma. J Ultrasound Med 2024; 43: 2269-2280.
doi: 10.1002/jum.16559 |
| [123] |
Jiang D , Ren J , Qian Y , Gu Y , Wang R , Yu H , et al . Prediction models after hepatectomy for hepatocellular carcinoma-based ultrasonic radiomics: an observational study. European journal of medical research 2025; 30: 722.
doi: 10.1186/s40001-025-02977-7 |
| [124] |
Liu D , Liu F , Xie X , Su L , Liu M , Xie X , et al . Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur Radiol 2020; 30: 2365-2376.
doi: 10.1007/s00330-019-06553-6 |
| [125] | Jin J , Yao Z , Zhang T , Zeng J , Wu L , Wu M , et al . Deep learning radiomics model accurately predicts hepatocellular carcinoma occurrence in chronic hepatitis B patients: a five-year follow-up. American journal of cancer research 2021; 11: 576-589 |
| [126] | Došilović FK , Brčić M , Hlupić N , editors. Explainable artificial intelligence: a survey. 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) 2018 21-25 May 2018. |
| [127] | Lambin P , Woodruff HC , Mali SA , Zhong X , Kuang S , Lavrova E , et al . Radiomics quality score 2. 0: towards radiomics readiness levels and clinical translation for personalized medicine. Nature Reviews Clinical Oncology 2025; 22: 831-846 |
| [1] | Hu Xuelin, Zhu Ye, Zhang Zisang, Quan Yuanting, Chen Wenwen, Chen Leichong, Xu Guangyu, Qin Luning, Xie Mingxing, Zhang Li. Explainable Artificial Intelligence in Echocardiography [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 409-425. |
| [2] | Guan Xin, Hu Xinyuan, Han Hong, Zhang Dezhi, Xu Huixiong. The Evolving Application of Ultrasound in the Precision Management of Small Hepatocellular Carcinoma [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 375-387. |
| [3] | Li Yanran, Cui Yuanjie, Wu Qingqing, Zhang Na. Current Applications of Artificial Intelligence in Obstetric Ultrasound [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 449-456. |
| [4] | Yu Xiao jie, Song Zheng lai, Chang Xue yong, Yu Jie, Liang Ping. Artificial Intelligence in Ultrasound Diagnosis of Liver Nodules: A Comprehensive Review of B-Mode and Contrast-enhanced Applications [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 326-346. |
| [5] | Jin Tong, Yu Xiaohu, Ai Zheng, Guo Hongcheng. Artificial Intelligence in Ultrasound Imaging: A Review of Progress from Machine Learning to Large Language Model [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 483-496. |
| [6] | Xiang Xi, Yang Yujia, Wang Liyun, Qiu Li. Advances and Applications in Dermatological Ultrasound [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(4): 457-466. |
| [7] | Zhang Simin, Zhou Changyu, Shi Xianquan, Huang Lizhen. Predictive Value of AIP and AGR for Non-alcoholic Fatty Liver Disease and Significant Liver Fibrosis [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(2): 197-206. |
| [8] | Liu Xinyu, Yuan Yinuo, Meng Yao, Zhang Wenjing, Xu Huimin, Zhang Jinrui, Liu Cun. Glomus Tumor of the Female Vulva: A Case Report and Review [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(2): 219-223. |
| [9] | Wang Yixuan, Jin Lin, Chen Jianxiong, Yang Huixian, Shen Cuiqin, Xu Wenzhe, Shen Yuzhou, Huang Jun, Sun Liwan, Du Lianfang, Wang Bei, Li Fan, Li Zhaojun. Is the Adventitial Vasa Vasorum in Vulnerable Carotid Plaques Increased or Decreased? [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(1): 56-64. |
| [10] | Sekarsari Damayanti, Gema Ramadhan Muhammad, Rinaldhy Kshetra, Prihartono Joedo. Biliary Atresia Treatment: Can Shear Wave Elastography Predict Kasai Procedure Outcomes? [J]. Advanced Ultrasound in Diagnosis and Therapy, 2025, 9(1): 84-91. |
| [11] | Mohammed Amr, Tahmasebi Aylin, Kim Sooji, Alnoury Mostafa, E. Wessner Corinne, Siu Xiao Tania, W. Gould Sharon, A. May Lauren, Kecskemethy Heidi, T. Saul David, R. Eisenbrey John. Evaluation of Liver Fibrosis on Grayscale Ultrasound in a Pediatric Population Using a Cloud-based Transfer Learning Artificial Intelligence Platform [J]. Advanced Ultrasound in Diagnosis and Therapy, 2024, 8(4): 242-249. |
| [12] | Liyun Xue, PhD, Hui Feng, MD, Fankun Meng, MD, Ying Zheng, MD, Guangwen Cheng, PhD, Yao Zhang, MD, Zhiyong Yin, MD, Jing Wu, MD, Jiabao Zhu, MD, Xueqi Li, MD, Jie Yu, PhD, Ping Liang, PhD, Hong Ding, PhD. Ultrasound-based Dual Elastography for Evaluating the Severity of Drug-induced Liver Injury: One Step Closer to Pathology [J]. Advanced Ultrasound in Diagnosis and Therapy, 2024, 8(3): 106-115. |
| [13] | Yue Tian, MB, Yanan Zhao, MD, Yiran Huang, MB. The Diagnostic Value of Real-time Shear Wave Ultrasound Elastography in the Differentiation of Hepatic Hemangioma and Hepatocellular Carcinoma [J]. Advanced Ultrasound in Diagnosis and Therapy, 2024, 8(3): 124-129. |
| [14] | Yuhang Zheng, BS, Jianqiao Zhou, MD. Deep Learning in Ultrasound Localization Microscopy [J]. Advanced Ultrasound in Diagnosis and Therapy, 2024, 8(3): 86-92. |
| [15] | Raymond Sutjiadi, MS, Siti Sendari, PhD, Heru Wahyu Herwanto, PhD, Yosi Kristian, PhD. Deep Learning for Segmentation and Classification in Mammograms for Breast Cancer Detection: A Systematic Literature Review [J]. Advanced Ultrasound in Diagnosis and Therapy, 2024, 8(3): 94-105. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
Share: WeChat
Copyright ©2018 Advanced Ultrasound in Diagnosis and Therapy
|
 Advanced Ultrasound in Diagnosis and Therapy (AUDT)
is licensed under a Creative Commons Attribution 4.0 International License.
Advanced Ultrasound in Diagnosis and Therapy (AUDT)
is licensed under a Creative Commons Attribution 4.0 International License.

