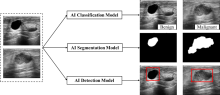
| [1] |
Reddy UM, Filly RA, Copel JA. Prenatal imaging: ultrasonography and magnetic resonance imaging. Obstet Gynecol 2008; 112:145-157.
doi: 10.1097/01.AOG.0000318871.95090.d9 pmid: 18591320 |
| [2] |
Anas EMA, Seitel A, Rasoulian A, John PS, Pichora D, Darras K, et al. Bone enhancement in ultrasound using local spectrum variations for guiding percutaneous scaphoid fracture fixation procedures. Int J CARS 2015; 10:959-969.
doi: 10.1007/s11548-015-1181-6 |
| [3] |
Noble JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans Med Imaging 2006; 25:987-1010.
doi: 10.1109/TMI.2006.877092 |
| [4] |
Pereira F, Bueno A, Rodriguez A, Perrin D, Marx G, Cardinale M, et al. Automated detection of coarctation of aorta in neonates from two dimensional echocardiograms. J Med Imaging 2017; 4:014502.
doi: 10.1117/1.JMI.4.1.014502 |
| [5] |
Chen C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. Journal of the American Society for Information Science and Technology 2006; 57:359-377.
doi: 10.1002/(ISSN)1532-2890 |
| [6] |
Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data Brief 2020; 28:104863.
doi: 10.1016/j.dib.2019.104863 |
| [7] |
Cao Z, Duan L, Yang G, Yue T, Chen Q. An experimental study on breast lesion detection and classification from ultrasound images using deep learning architectures. BMC medical imaging 2019; 19:1-9.
doi: 10.1186/s12880-018-0301-5 |
| [8] |
Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Deep learning approaches for data augmentation and classification of breast masses using ultrasound images. Int. J. Adv. Comput. Sci. Appl 2019; 10:1-11.
doi: 10.3390/app10010001 |
| [9] | Han X, Wang J, Zhou W, Chang C, Ying S, Shi J. Deep doubly supervised transfer network for diagnosis of breast cancer with imbalanced ultrasound imaging modalities//Medical Image Computing and Computer Assisted Intervention-MICCAI 2020: 23rd International Conference, Lima, Peru, October 4-8, 2020, Proceedings, Part VI 23. Springer International Publishing; 2020:141-149. |
| [10] |
Zhou Y, Chen H, Li Y, Liu Q, Xu X, Wang S, et al. Multi-task learning for segmentation and classification of tumors in 3D automated breast ultrasound images. Medical Image Analysis 2021; 70:101918.
doi: 10.1016/j.media.2020.101918 |
| [11] | Badawy SM, Mohamed AENA, Hefnawy AA, Zidan HE, GadAllah MT, El-Banby GM. Automatic semantic segmentation of breast tumors in ultrasound images based on combining fuzzy logic and deep learning—a feasibility study. PLoS One 2021; 16:e0251899. |
| [12] |
Bourouis S, Band SS, Mosavi A, Agrawal S, Ahila A, Poongodi M, et al. Meta-heuristic algorithm-tuned neural network for breast cancer diagnosis using ultrasound images. Frontiers in Oncology 2022; 12:834028.
doi: 10.3389/fonc.2022.834028 |
| [13] |
Jabeen K, Khan MA, Alhaisoni M, Tariq U, Zhang YD, Hamza A, et al. Breast cancer classification from ultrasound images using probability-based optimal deep learning feature fusion. Sensors 2022; 22:807.
doi: 10.3390/s22030807 |
| [14] |
Ragab M, Albukhari A, Alyami J, Mansour RF. Ensemble deep-learning-enabled clinical decision support system for breast cancer diagnosis and classification on ultrasound images. Biology 2022; 11:439.
doi: 10.3390/biology11030439 |
| [15] | Gheflati B, Rivaz H. Vision transformers for classification of breast ultrasound images. Annu Int Conf IEEE Eng Med Biol Soc 2022:480-483. |
| [16] |
Ayana G, Park J, Jeong JW, Choe S. A novel multistage transfer learning for ultrasound breast cancer image classification. Diagnostics 2022; 12:135.
doi: 10.3390/diagnostics12010135 |
| [17] |
Liu T, Guo Q, Lian C, Ren X, Liang S, Yu J, et al. Automated detection and classification of thyroid nodules in ultrasound images using clinical-knowledge-guided convolutional neural networks. Medical image analysis 2019; 58:101555.
doi: 10.1016/j.media.2019.101555 |
| [18] |
Kuo CC, Chang CM, Liu KT, Lin WK, Chiang HY, Chung CW, et al. Automation of the kidney function prediction and classification through ultrasound-based kidney imaging using deep learning. NPJ digital medicine 2019; 2:29.
doi: 10.1038/s41746-019-0104-2 |
| [19] |
Roy S, Menapace W, Oei S, Luijten B, Fini E, Saltori. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans Med Imaging 2020; 39:2676-2687.
doi: 10.1109/TMI.42 |
| [20] |
Xie HN, Wang N, He M, Zhang LH, Cai HM, Xian B J, et al. Using deep-learning algorithms to classify fetal brain ultrasound images as normal or abnormal. Ultrasound Obstet Gynecol 2020; 56:579-587.
doi: 10.1002/uog.21967 pmid: 31909548 |
| [21] | Skandha SS, Gupta SK, Saba L, Koppula VK, Johri AM, Khanna NN, et al. 3-D optimized classification and characterization artificial intelligence paradigm for cardiovascular/stroke risk stratification using carotid ultrasound-based delineated plaque: AtheromaticTM 2.0. Comput Biol Med 2020;125103958. |
| [22] | Chen H, Zheng Y, Park JH, Heng PA, Zhou SK, et al. Iterative multi-domain regularized deep learning for anatomical structure detection and segmentation from ultrasound images. IEEE Transactions on Medical Imaging 2019; 38:2514-2524. |
| [23] |
Cui W, Meng D, Lu K, Wang L, Zhou X, Li Y, et al. Automatic segmentation of ultrasound images using SegNet and local Nakagami distribution fitting model. Biomedical Signal Processing and Control 2023; 81:104431.
doi: 10.1016/j.bspc.2022.104431 |
| [24] | Dangoury S, Sadik M, Alali A, Fail A, Khoury R, Nassar J. V-net performances for 2D ultrasound image segmentation. IEEE 18th International Colloquium on Signal Processing & Applications (CSPA) 2022:96-100. |
| [25] | Yap MH, Pons G, Martí J, Ganau S, Sentís M, Zwiggelaar R, et al. Automated breast ultrasound lesions detection using convolutional neural networks. IIEEE J Biomed Health Inform 2018; 22:1218-1226. |
| [26] | Sharifzadeh M, Benali H, Rivaz H. Shift-invariant segmentation in breast ultrasound images. IEEE International Ultrasonics Symposium (IUS) 2021:1-4. |
| [27] |
Gare GR, Li J, Joshi R, Patel S, Kumar V, Shah P, et al. W-Net: Dense and diagnostic semantic segmentation of subcutaneous and breast tissue in ultrasound images by incorporating ultrasound RF waveform data. Medical Image Analysis 2022; 76:102326.
doi: 10.1016/j.media.2021.102326 |
| [28] |
Yin S, Peng Q, Li H, Gao X, Liu L, Feng S, et al. Automatic kidney segmentation in ultrasound images using subsequent boundary distance regression and pixelwise classification networks. Medical Image Analysis 2020; 60:101602.
doi: 10.1016/j.media.2019.101602 |
| [29] | Torres HR, Queirós S, Morais P, Fonseca JC, Vilaça JL, Oliveira C, et al. Kidney segmentation in 3-D ultrasound images using a fast phase-based approach. IEEE Trans Ultrason Ferroelectr Freq Control 2020;68:1521-1531. |
| [30] |
Chen G, Dai Y, Li R, Wang J, Wu X, Zhang L, et al. SDFNet: Automatic segmentation of kidney ultrasound images using multi-scale low-level structural feature. Expert Systems with Applications 2021; 185:115619.
doi: 10.1016/j.eswa.2021.115619 |
| [31] |
Valente S, Ferreira HA, Silva M, Oliveira R, Santos J, Gomes P, et al. A deep learning method for kidney segmentation in 2D ultrasound images. Annu Int Conf IEEE Eng Med Biol Soc 2022: 2022:3911-3914.
doi: 10.1109/EMBC48229.2022.9871748 pmid: 36086291 |
| [32] | Leclerc S, Smistad E, Pedrosa J, Østvik A, Cervenansky F, Espinosa F, et al. Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE Trans Med Imaging 2019;38:2198-2210. |
| [33] |
Pu B, Wang Q, Zhang Y, Zhao H, Liu Y, Chen X, et al. MobileUNet-FPN: A semantic segmentation model for fetal ultrasound four-chamber segmentation in edge computing environments. IEEE J Biomed Health Inform 2022; 26:5540-5550.
doi: 10.1109/JBHI.2022.3182722 |
| [34] |
Ma J, Wu F, Jiang T, Zhao Q, Kong D. Ultrasound image-based thyroid nodule automatic segmentation using convolutional neural networks. Int J Comput Assist Radiol Surg 2018; 13:1685-1697.
doi: 10.1007/s11548-018-1815-6 |
| [35] |
Li H, Fang J, Liu S, Liang X, Yang X, Mai Z, et al. Cr-UNet: a composite network for ovary and follicle segmentation in ultrasound images. IEEE J Biomed Health Inform 2019; 24:974-983.
doi: 10.1109/JBHI.6221020 |
| [36] | Qiu Z, Qin Y, Duan X, Shi Y, Yang X, Qin J, et al. Automatic mouse embryo brain ventricle & body segmentation and mutant classification from ultrasound data using deep learning. IEEE International Ultrasonics Symposium (IUS) 2019:12-15. |
| [37] |
Kim J, Kim HJ, Kim C, Lee JH, Kim KW, Park YM, et al. Weakly-supervised deep learning for ultrasound diagnosis of breast cancer. Scientific Reports 2021; 11:24382.
doi: 10.1038/s41598-021-03806-7 pmid: 34934144 |
| [38] |
Shen Y, Shamout FE, Oliver JR, Witowski J, Kannan K, Park J, et al. Artificial intelligence system reduces false-positive findings in the interpretation of breast ultrasound exams. Nature communications 2021; 12:5645.
doi: 10.1038/s41467-021-26023-2 pmid: 34561440 |
| [39] |
Niu S, Huang J, Li J, Liu X, Wang D, Zhang RF, et al. Application of ultrasound artificial intelligence in the differential diagnosis between benign and malignant breast lesions of BI-RADS 4A. BMC cancer 2020; 20:1-7.
doi: 10.1186/s12885-019-6169-0 |
| [40] |
Zhang X, Lin X, Zhang Z, Dong LC, Sun XL, Sun DS, et al. Artificial intelligence medical ultrasound equipment: application of breast lesions detection. Ultrasonic Imaging 2020; 42:191-202.
doi: 10.1177/0161734620928453 |
| [41] |
Qian X, Pei J, Zheng H, Xie X, Yan L, Zhang H, et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nat Biomed Eng 2021; 5:522-532.
doi: 10.1038/s41551-021-00711-2 pmid: 33875840 |
| [42] |
Baloescu C, Toporek G, Kim S, McNamara K, Liu R, Shaw M, et al. Automated lung ultrasound B-line assessment using a deep learning algorithm. IEEE Trans Ultrason Ferroelectr Freq Control 2020; 67:2312-2320.
doi: 10.1109/TUFFC.58 |
| [43] | Diaz-Escobar J, Ordóñez-Guillén NE, Villarreal-Reyes S, Galaviz-Mosqueda A, Kober V, Rivera-Rodriguez R, et al. Deep-learning based detection of COVID-19 using lung ultrasound imagery. Plos one 2021; 16:e0255886. |
| [44] |
Fang X, Li W, Huang J, Li W, Feng Q, Han Y, et al. Ultrasound image intelligent diagnosis in community-acquired pneumonia of children using convolutional neural network based transfer learning. Front Pediatr 2022; 10:1063587.
doi: 10.3389/fped.2022.1063587 |
| [45] | Kulhare S, Zheng X, Mehanian C, Gregory C, Zhu M, Gregory K, et al. Ultrasound-based detection of lung abnormalities using single shot detection convolutional neural networks. In Simulation, Image Processing, and Ultrasound Systems for Assisted Diagnosis and Navigation: International Workshops 2018;pp:65-73. |
| [46] |
Abdel-Basset M, Hawash H, Alnowibet KA, Mohamed AW, Sallam KM. Interpretable deep learning for discriminating pneumonia from lung ultrasounds. Mathematics 2022; 10:4153.
doi: 10.3390/math10214153 |
| [47] |
Choi YJ, Baek JH, Park HS, Shim WH, Kim TY, Shong YK, et al. A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of thyroid nodules on ultrasound: initial clinical assessment. Thyroid 2017; 27:546-552.
doi: 10.1089/thy.2016.0372 pmid: 28071987 |
| [48] | Wei X, Zhu J, Zhang H, Gao H, Yu R, Liu Z, et al. Visual interpretability in computer-assisted diagnosis of thyroid nodules using ultrasound images. Med Sci Monit 2020; 26:e927007. |
| [49] |
Wang L, Yang S, Yang S, Zhao C, Tian G, Gao Y, et al. Automatic thyroid nodule recognition and diagnosis in ultrasound imaging with the YOLOv2 neural network. World J Surg Oncol 2019; 17:1-9.
doi: 10.1186/s12957-018-1541-0 |
| [50] |
Jassal K, Koohestani A, Kiu A, Strong A, Ravintharan N, Yeung M, et al. Artificial intelligence for pre-operative diagnosis of malignant thyroid nodules based on sonographic features and cytology category. World J Surg 2023; 47:330-339.
doi: 10.1007/s00268-022-06798-1 |
| [51] | Deng C, Han D, Feng M, Lv Z, Li D. Differential diagnostic value of the ResNet50, random forest, and DS ensemble models for papillary thyroid carcinoma and other thyroid nodules. J Int Med Res 2022;50. |
| [52] |
Fujioka T, Mori M, Kubota K, Kikuchi Y, Katsuta L, Adachi M, et al. Breast ultrasound image synthesis using deep convolutional generative adversarial networks. Diagnostics 2019; 9:176.
doi: 10.3390/diagnostics9040176 |
| [53] | Jarosik P, Lewandowski M, Klimonda Z, Byra M. Pixel-wise deep reinforcement learning approach for ultrasound image denoising. 2021 IEEE International Ultrasounds Symposium (IUS) 2021;1-4. |
| [54] |
Czajkowska J, Juszczyk J, Piejko L, Glenc-Ambroży M. High-frequency ultrasound dataset for deep learning-based image quality assessment. Sensors 2022; 22:1478.
doi: 10.3390/s22041478 |
| [55] |
Wang J, Yang X, Zhou B, Sohn JJ, Zhou J, Jacob JT, et al. Review of machine learning in lung ultrasound in COVID-19 pandemic. Journal of Imaging 2022; 8:65.
doi: 10.3390/jimaging8030065 |
| No related articles found! |
|
||